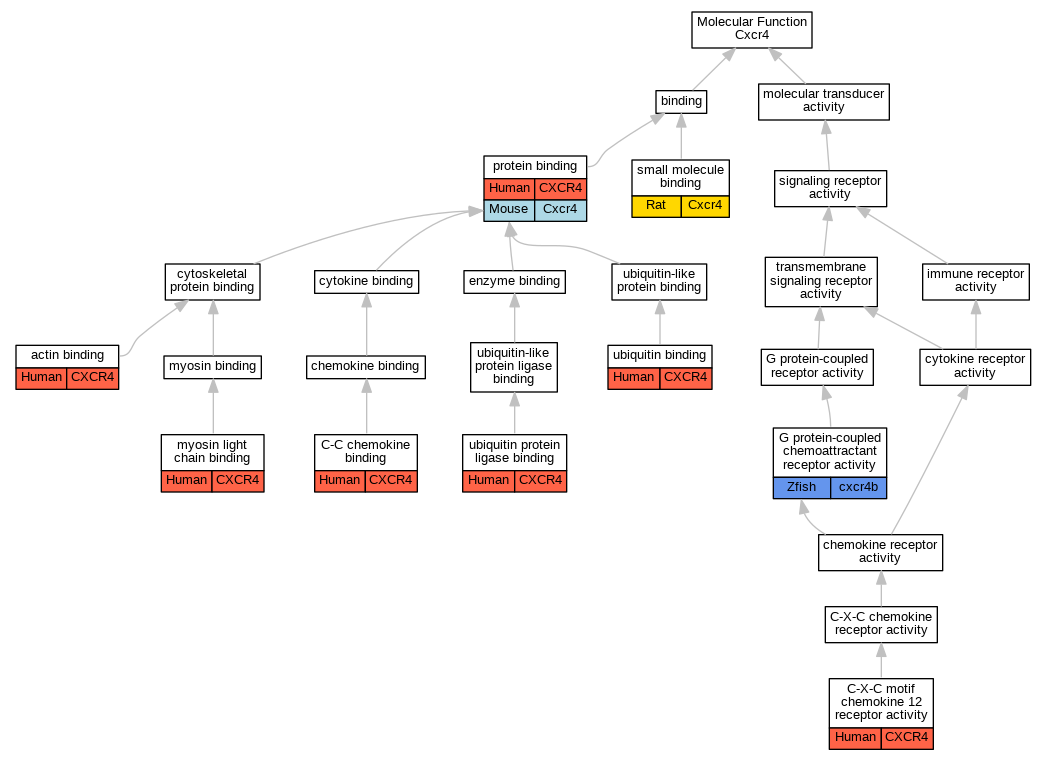
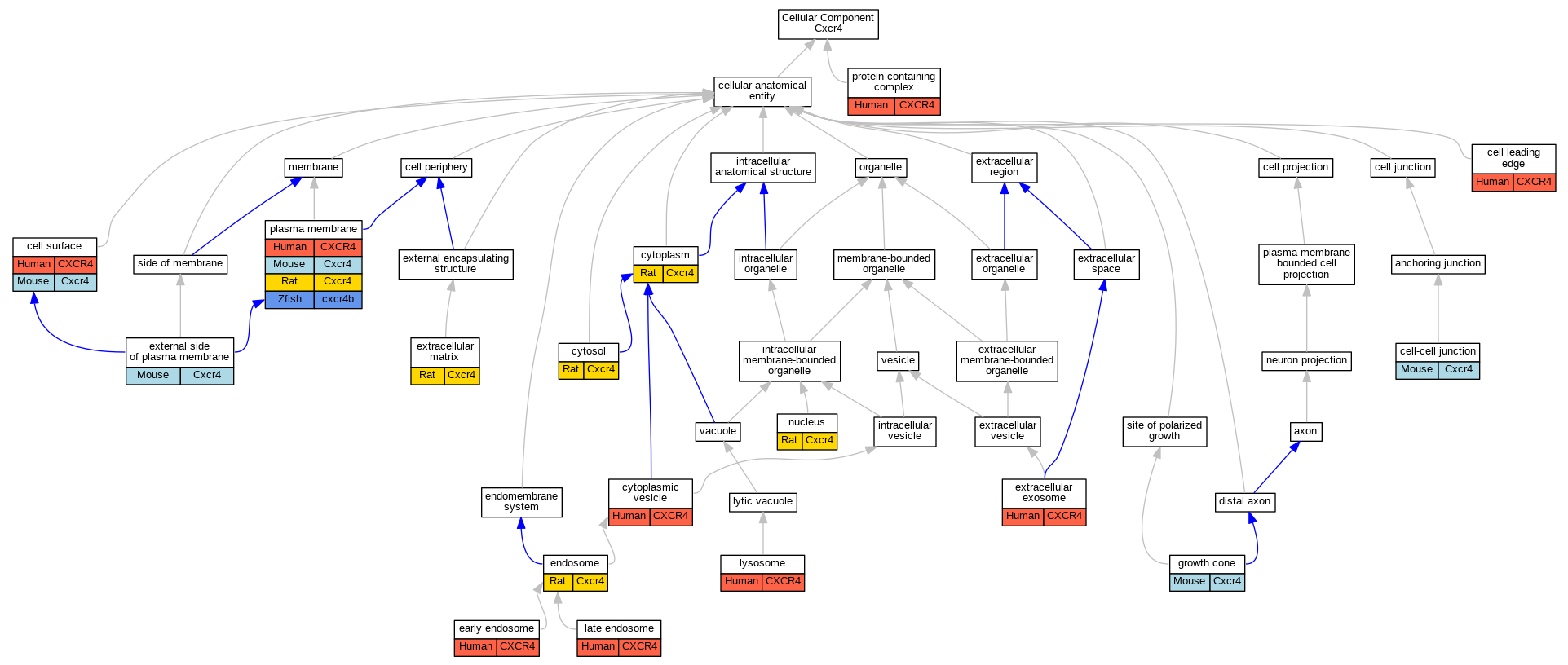
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003779 | actin binding | CXCR4 | IDA | | Human | PMID:12421915 |
| Molecular Function | GO:0019957 | C-C chemokine binding | CXCR4 | IPI | UniProtKB:Q98157 | Human | PMID:25612609 |
| Molecular Function | GO:0038147 | C-X-C motif chemokine 12 receptor activity | CXCR4 | IDA | | Human | PMID:28978524 |
| Molecular Function | GO:0001637 | G protein-coupled chemoattractant receptor activity | cxcr4b | IMP | | Zfish | PMID:15340012 |
| Molecular Function | GO:0001637 | G protein-coupled chemoattractant receptor activity | cxcr4b | IMP | | Zfish | PMID:15081362 |
| Molecular Function | GO:0032027 | myosin light chain binding | CXCR4 | IDA | | Human | PMID:12421915 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P32121 | Human | PMID:10644702 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P35579 | Human | PMID:12421915 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P51681 | Human | PMID:15660419 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q04900 | Human | PMID:17077324 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P51681 | Human | PMID:18632580 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:O60603 | Human | PMID:18765807 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P54578 | Human | PMID:19106094 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:19380869 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P04233 | Human | PMID:19665027 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q07266-1 | Human | PMID:20215400 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q16643 | Human | PMID:20215400 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:O60603 | Human | PMID:20802527 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:21730065 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P49682 | Human | PMID:23170857 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P32302 | Human | PMID:23773523 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q16643 | Human | PMID:23926103 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q04900 | Human | PMID:24094005 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:25775528 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:27331810 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:28862946 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:O15541 | Human | PMID:28978524 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:P25106 | Human | PMID:29386406 |
| Molecular Function | GO:0005515 | protein binding | CXCR4 | IPI | UniProtKB:Q96J02-2 | Human | PMID:34927784 |
| Molecular Function | GO:0005515 | protein binding | Cxcr4 | IPI | UniProtKB:Q9R0L9 | Mouse | MGI:MGI:3793396|PMID:18227060 |
| Molecular Function | GO:0005515 | protein binding | Cxcr4 | IPI | PR:Q8BGW2 | Mouse | PMID:31845480 |
| Molecular Function | GO:0036094 | small molecule binding | Cxcr4 | IMP | | Rat | PMID:21294880|RGD:6480473 |
| Molecular Function | GO:0043130 | ubiquitin binding | CXCR4 | IDA | | Human | PMID:20228059 |
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | CXCR4 | IPI | UniProtKB:Q96J02 | Human | PMID:19116316 |
| Cellular Component | GO:0031252 | cell leading edge | CXCR4 | IDA | | Human | PMID:12421915 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:10521508 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:10644702 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:18308860 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:19106094 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:19665027 |
| Cellular Component | GO:0009986 | cell surface | CXCR4 | IDA | | Human | PMID:23620790 |
| Cellular Component | GO:0009986 | cell surface | Cxcr4 | IDA | | Mouse | MGI:MGI:5448504|PMID:22265737 |
| Cellular Component | GO:0005911 | cell-cell junction | Cxcr4 | IDA | | Mouse | PMID:23793062 |
| Cellular Component | GO:0005737 | cytoplasm | Cxcr4 | IDA | | Rat | PMID:24924806|RGD:11352662 |
| Cellular Component | GO:0031410 | cytoplasmic vesicle | CXCR4 | IDA | | Human | PMID:10521508 |
| Cellular Component | GO:0031410 | cytoplasmic vesicle | CXCR4 | IDA | | Human | PMID:19106094 |
| Cellular Component | GO:0005829 | cytosol | Cxcr4 | IDA | | Rat | PMID:24912495|RGD:151893463 |
| Cellular Component | GO:0005769 | early endosome | CXCR4 | IDA | | Human | PMID:24790097 |
| Cellular Component | GO:0005769 | early endosome | CXCR4 | IDA | | Human | PMID:21540189 |
| Cellular Component | GO:0005768 | endosome | Cxcr4 | IDA | | Rat | PMID:16952464|RGD:1600771 |
| Cellular Component | GO:0009897 | external side of plasma membrane | Cxcr4 | IDA | | Mouse | PMID:19008373 |
| Cellular Component | GO:0070062 | extracellular exosome | CXCR4 | HDA | | Human | PMID:20458337 |
| Cellular Component | GO:0031012 | extracellular matrix | Cxcr4 | IDA | | Rat | PMID:24912495|RGD:151893463 |
| Cellular Component | GO:0030426 | growth cone | Cxcr4 | IDA | | Mouse | PMID:16129397 |
| Cellular Component | GO:0005770 | late endosome | CXCR4 | IDA | | Human | PMID:21540189 |
| Cellular Component | GO:0005764 | lysosome | CXCR4 | IDA | | Human | PMID:24790097 |
| Cellular Component | GO:0005764 | lysosome | CXCR4 | IDA | | Human | PMID:21540189 |
| Cellular Component | GO:0005634 | nucleus | Cxcr4 | IDA | | Rat | PMID:29550796|RGD:13792610 |
| Cellular Component | GO:0005886 | plasma membrane | CXCR4 | IDA | | Human | PMID:19665027 |
| Cellular Component | GO:0005886 | plasma membrane | CXCR4 | IDA | | Human | PMID:21540189 |
| Cellular Component | GO:0005886 | plasma membrane | Cxcr4 | IDA | | Mouse | PMID:20376365 |
| Cellular Component | GO:0005886 | plasma membrane | Cxcr4 | IDA | | Rat | PMID:17046839|RGD:2306583 |
| Cellular Component | GO:0005886 | plasma membrane | Cxcr4 | IDA | | Rat | PMID:24912495|RGD:151893463 |
| Cellular Component | GO:0005886 | plasma membrane | cxcr4b | IDA | | Zfish | PMID:20592249 |
| Cellular Component | GO:0032991 | protein-containing complex | CXCR4 | IDA | | Human | PMID:19665027 |
| Cellular Component | GO:0032991 | protein-containing complex | CXCR4 | IMP | | Human | PMID:25612609 |
| Biological Process | GO:0001667 | ameboidal-type cell migration | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:16678780 |
| Biological Process | GO:0009887 | animal organ morphogenesis | cxcr4b | IMP | | Zfish | PMID:12444253 |
| Biological Process | GO:0035904 | aorta development | Cxcr4 | IMP | | Mouse | PMID:25807483 |
| Biological Process | GO:0007411 | axon guidance | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-051006-4 | Zfish | PMID:19595689 |
| Biological Process | GO:0007411 | axon guidance | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:17537794 |
| Biological Process | GO:0007411 | axon guidance | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:15716407 |
| Biological Process | GO:0007411 | axon guidance | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:15097993 |
| Biological Process | GO:0007411 | axon guidance | cxcr4b | IGI | ZFIN:ZDB-MRPHLNO-050318-6,ZFIN:ZDB-MRPHLNO-050318-7 | Zfish | PMID:15716407 |
| Biological Process | GO:0043534 | blood vessel endothelial cell migration | Cxcr4 | IMP | MGI:MGI:1934442 | Mouse | PMID:15626744 |
| Biological Process | GO:0007420 | brain development | Cxcr4 | IMP | MGI:MGI:1934407 | Mouse | PMID:12183377 |
| Biological Process | GO:0001569 | branching involved in blood vessel morphogenesis | Cxcr4 | IMP | MGI:MGI:1934442 | Mouse | PMID:15626744 |
| Biological Process | GO:0019722 | calcium-mediated signaling | CXCR4 | IMP | | Human | PMID:19064997 |
| Biological Process | GO:0060048 | cardiac muscle contraction | Cxcr4 | IMP | | Rat | PMID:17010372|RGD:1600772 |
| Biological Process | GO:0016477 | cell migration | Cxcr4 | IMP | | Rat | PMID:15358596|RGD:1600768 |
| Biological Process | GO:0016477 | cell migration | cxcr4b | IMP | | Zfish | PMID:12444253 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-081114-3 | Zfish | PMID:18579679 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-070427-1 | Zfish | PMID:23884240 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-081114-4 | Zfish | PMID:18579679 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | cxcr4a | IMP | ZFIN:ZDB-GENO-130925-15 | Zfish | PMID:23884240 |
| Biological Process | GO:0002042 | cell migration involved in sprouting angiogenesis | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-110429-3 | Zfish | PMID:21429985 |
| Biological Process | GO:0002042 | cell migration involved in sprouting angiogenesis | cxcr4a | IGI | ZFIN:ZDB-MRPHLNO-110429-2,ZFIN:ZDB-MRPHLNO-110429-3 | Zfish | PMID:21429985 |
| Biological Process | GO:0000902 | cell morphogenesis | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-081114-3 | Zfish | PMID:18579679 |
| Biological Process | GO:0071345 | cellular response to cytokine stimulus | CXCR4 | IDA | | Human | PMID:21540189 |
| Biological Process | GO:0071345 | cellular response to cytokine stimulus | Cxcr4 | IDA | | Mouse | PMID:8962122 |
| Biological Process | GO:0071417 | cellular response to organonitrogen compound | Cxcr4 | IPI | CHEBI:125354 | Rat | PMID:24912495|RGD:151893463 |
| Biological Process | GO:0071466 | cellular response to xenobiotic stimulus | Cxcr4 | IEP | | Rat | PMID:20613717|RGD:11352688 |
| Biological Process | GO:0060271 | cilium assembly | cxcr4a | IGI | ZFIN:ZDB-GENE-980526-176 | Zfish | PMID:31430272 |
| Biological Process | GO:0060271 | cilium assembly | cxcr4a | IMP | ZFIN:ZDB-GENO-200330-2 | Zfish | PMID:31430272 |
| Biological Process | GO:0060271 | cilium assembly | cxcr4a | IGI | ZFIN:ZDB-GENE-060929-1178 | Zfish | PMID:31430272 |
| Biological Process | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | CXCR4 | IMP | | Human | PMID:24912431 |
| Biological Process | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | CXCR4 | IDA | | Human | PMID:28978524 |
| Biological Process | GO:0038160 | CXCL12-activated CXCR4 signaling pathway | Cxcr4 | IDA | | Mouse | PMID:31845480 |
| Biological Process | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | Cxcr4 | IMP | | Rat | PMID:27760212|RGD:12910551 |
| Biological Process | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain | Cxcr4 | IMP | | Rat | PMID:28108674|RGD:13463594 |
| Biological Process | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | Cxcr4 | IMP | | Rat | PMID:27760212|RGD:12910551 |
| Biological Process | GO:0007368 | determination of left/right symmetry | cxcr4b | IMP | ZFIN:ZDB-GENO-200709-5 | Zfish | PMID:31371827 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | cxcr4a | IGI | ZFIN:ZDB-GENE-031009-1 | Zfish | PMID:25607881 |
| Biological Process | GO:0045446 | endothelial cell differentiation | Cxcr4 | IMP | | Rat | PMID:24955809|RGD:11352686 |
| Biological Process | GO:0061154 | endothelial tube morphogenesis | Cxcr4 | IMP | | Rat | PMID:24955809|RGD:11352686 |
| Biological Process | GO:0002064 | epithelial cell development | Cxcr4 | IMP | | Rat | PMID:15358596|RGD:1600768 |
| Biological Process | GO:0030900 | forebrain development | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-051006-4 | Zfish | PMID:19595689 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | CXCR4 | IDA | | Human | PMID:10644702 |
| Biological Process | GO:0000082 | G1/S transition of mitotic cell cycle | cxcr4a | IMP | ZFIN:ZDB-GENO-200330-2 | Zfish | PMID:31430272 |
| Biological Process | GO:0000082 | G1/S transition of mitotic cell cycle | cxcr4a | IGI | ZFIN:ZDB-GENE-980526-176 | Zfish | PMID:31430272 |
| Biological Process | GO:0048699 | generation of neurons | Cxcr4 | IEP | | Mouse | MGI:MGI:4943615|PMID:20882566 |
| Biological Process | GO:0007281 | germ cell development | Cxcr4 | IMP | MGI:MGI:1934407 | Mouse | PMID:12900445 |
| Biological Process | GO:0007281 | germ cell development | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:17517144 |
| Biological Process | GO:0008354 | germ cell migration | Cxcr4 | IMP | MGI:MGI:1934407 | Mouse | PMID:12900445 |
| Biological Process | GO:0008354 | germ cell migration | cxcr4b | IMP | | Zfish | PMID:15340012 |
| Biological Process | GO:0008354 | germ cell migration | cxcr4b | IMP | | Zfish | PMID:15068797 |
| Biological Process | GO:0035701 | hematopoietic stem cell migration | Cxcr4 | IMP | MGI:MGI:3525214 | Mouse | PMID:20847314 |
| Biological Process | GO:0050930 | induction of positive chemotaxis | cxcr4b | IMP | | Zfish | PMID:15068797 |
| Biological Process | GO:0001822 | kidney development | Cxcr4 | IMP | MGI:MGI:5297422 | Mouse | PMID:27002738 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcr4a | IGI | ZFIN:ZDB-GENE-980526-176 | Zfish | PMID:31430272 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcr4a | IMP | ZFIN:ZDB-GENO-200330-2 | Zfish | PMID:31430272 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcr4b | IMP | | Zfish | PMID:31371827 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcr4b | IMP | ZFIN:ZDB-GENO-200709-9 | Zfish | PMID:31371827 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | cxcr4b | IMP | ZFIN:ZDB-GENO-200709-8 | Zfish | PMID:31371827 |
| Biological Process | GO:0051674 | localization of cell | cxcr4b | IMP | ZFIN:ZDB-GENO-200709-8 | Zfish | PMID:31371827 |
| Biological Process | GO:0001946 | lymphangiogenesis | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-120507-1 | Zfish | PMID:24903752 |
| Biological Process | GO:0060231 | mesenchymal to epithelial transition | cxcr4a | IGI | ZFIN:ZDB-GENE-030710-1 | Zfish | PMID:28701377 |
| Biological Process | GO:0060231 | mesenchymal to epithelial transition | cxcr4a | IGI | ZFIN:ZDB-GENE-020708-3 | Zfish | PMID:28701377 |
| Biological Process | GO:0060231 | mesenchymal to epithelial transition | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-071022-1 | Zfish | PMID:28701377 |
| Biological Process | GO:0008045 | motor neuron axon guidance | Cxcr4 | IMP | MGI:MGI:1934442 | Mouse | PMID:16129397 |
| Biological Process | GO:0008045 | motor neuron axon guidance | Cxcr4 | IMP | MGI:MGI:3777572 | Mouse | PMID:18031680 |
| Biological Process | GO:0043217 | myelin maintenance | Cxcr4 | IMP | | Mouse | MGI:MGI:4458974|PMID:20534485 |
| Biological Process | GO:0030239 | myofibril assembly | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-071022-1 | Zfish | PMID:17517144 |
| Biological Process | GO:0090024 | negative regulation of neutrophil chemotaxis | cxcr4b | IMP | ZFIN:ZDB-GENO-110209-3 | Zfish | PMID:20592249 |
| Biological Process | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development | cxcr4a | IGI | ZFIN:ZDB-GENE-031009-1 | Zfish | PMID:25607881 |
| Biological Process | GO:0061351 | neural precursor cell proliferation | Cxcr4 | IEP | | Mouse | MGI:MGI:4943615|PMID:20882566 |
| Biological Process | GO:0022008 | neurogenesis | Cxcr4 | IMP | | Rat | PMID:18434527|RGD:11352690 |
| Biological Process | GO:0001764 | neuron migration | Cxcr4 | IDA | | Mouse | PMID:12900445 |
| Biological Process | GO:0001764 | neuron migration | Cxcr4 | IDA | | Rat | PMID:12401342|RGD:1358280 |
| Biological Process | GO:0001764 | neuron migration | Cxcr4 | IMP | | Rat | PMID:15048928|RGD:1600767 |
| Biological Process | GO:0001764 | neuron migration | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:23382464 |
| Biological Process | GO:0001764 | neuron migration | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:16129396 |
| Biological Process | GO:0001764 | neuron migration | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-051006-4 | Zfish | PMID:19595689 |
| Biological Process | GO:0008038 | neuron recognition | Cxcr4 | IEP | | Rat | PMID:12431218|RGD:632605 |
| Biological Process | GO:0030316 | osteoclast differentiation | Cxcr4 | HEP | | Rat | PMID:16304045|RGD:151665775 |
| Biological Process | GO:0050918 | positive chemotaxis | cxcr4b | IMP | | Zfish | PMID:15340012 |
| Biological Process | GO:0090280 | positive regulation of calcium ion import | Cxcr4 | IDA | | Mouse | PMID:8962122 |
| Biological Process | GO:0030335 | positive regulation of cell migration | CXCR4 | IMP | | Human | PMID:20023705 |
| Biological Process | GO:0030335 | positive regulation of cell migration | Cxcr4 | IMP | | Rat | PMID:24924806|RGD:11352662 |
| Biological Process | GO:0050921 | positive regulation of chemotaxis | Cxcr4 | IMP | | Rat | PMID:30221695|RGD:14348966 |
| Biological Process | GO:0120162 | positive regulation of cold-induced thermogenesis | Cxcr4 | IMP | | Mouse | MGI:MGI:5614545|PMID:25016030 |
| Biological Process | GO:1903861 | positive regulation of dendrite extension | Cxcr4 | IMP | | Rat | PMID:28104461|RGD:13463105 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | Cxcr4 | IMP | | Mouse | PMID:20376365 |
| Biological Process | GO:2000448 | positive regulation of macrophage migration inhibitory factor signaling pathway | CXCR4 | IMP | | Human | PMID:19665027 |
| Biological Process | GO:1905322 | positive regulation of mesenchymal stem cell migration | Cxcr4 | IMP | | Rat | PMID:24924806|RGD:11352662 |
| Biological Process | GO:0050769 | positive regulation of neurogenesis | Cxcr4 | IMP | | Rat | PMID:28104461|RGD:13463105 |
| Biological Process | GO:0048714 | positive regulation of oligodendrocyte differentiation | Cxcr4 | IMP | | Mouse | MGI:MGI:4458974|PMID:20534485 |
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | Cxcr4 | IMP | MGI:MGI:3525214 | Mouse | PMID:20847314 |
| Biological Process | GO:0035470 | positive regulation of vascular wound healing | Cxcr4 | IMP | | Rat | PMID:24955809|RGD:11352686 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IGI | ZFIN:ZDB-MRPHLNO-081217-1,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-081217-1 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IGI | ZFIN:ZDB-GENO-071022-3,ZFIN:ZDB-MRPHLNO-100608-9 | Zfish | PMID:20308561 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IGI | ZFIN:ZDB-GENO-030312-1,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17570670 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IGI | ZFIN:ZDB-GENO-140512-3,ZFIN:ZDB-MRPHLNO-070126-7,ZFIN:ZDB-MRPHLNO-120123-1,ZFIN:ZDB-MRPHLNO-120123-2 | Zfish | PMID:24201188 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IGI | ZFIN:ZDB-MRPHLNO-050318-7,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-050318-7 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:17570670 |
| Biological Process | GO:0030516 | regulation of axon extension | cxcr4b | IDA | | Zfish | PMID:15097993 |
| Biological Process | GO:0030516 | regulation of axon extension | cxcr4b | IMP | ZFIN:ZDB-GENO-030312-1 | Zfish | PMID:15081362 |
| Biological Process | GO:0051924 | regulation of calcium ion transport | Cxcr4 | IMP | | Rat | PMID:12864967|RGD:1299272 |
| Biological Process | GO:0030155 | regulation of cell adhesion | CXCR4 | IDA | | Human | PMID:19703720 |
| Biological Process | GO:0030334 | regulation of cell migration | Cxcr4 | IMP | MGI:MGI:1934407 | Mouse | PMID:12900445 |
| Biological Process | GO:0030334 | regulation of cell migration | Cxcr4 | IGI | MGI:MGI:108088 | Mouse | PMID:16166380 |
| Biological Process | GO:0050920 | regulation of chemotaxis | CXCR4 | IMP | | Human | PMID:19106094 |
| Biological Process | GO:0050920 | regulation of chemotaxis | Cxcr4 | IMP | | Rat | PMID:15358596|RGD:1600768 |
| Biological Process | GO:0106014 | regulation of inflammatory response to wounding | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:28220184 |
| Biological Process | GO:0051453 | regulation of intracellular pH | cxcr4b | IMP | ZFIN:ZDB-MRPHLNO-050318-7 | Zfish | PMID:25843033 |
| Biological Process | GO:2001222 | regulation of neuron migration | Cxcr4 | IMP | MGI:MGI:1934407 | Mouse | PMID:12183377 |
| Biological Process | GO:0090022 | regulation of neutrophil chemotaxis | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:28220184 |
| Biological Process | GO:0043067 | regulation of programmed cell death | Cxcr4 | IMP | | Rat | PMID:14550764|RGD:1600766 |
| Biological Process | GO:0050792 | regulation of viral process | Cxcr4 | IDA | | Rat | PMID:9060691|RGD:11352687 |
| Biological Process | GO:0014823 | response to activity | Cxcr4 | IEP | | Rat | PMID:22140095|RGD:11352685 |
| Biological Process | GO:0009617 | response to bacterium | cxcr4b | IMP | ZFIN:ZDB-GENO-170614-1 | Zfish | PMID:28332618 |
| Biological Process | GO:0001666 | response to hypoxia | CXCR4 | IEP | | Human | PMID:15174142 |
| Biological Process | GO:0032496 | response to lipopolysaccharide | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:19110060 |
| Biological Process | GO:0043278 | response to morphine | Cxcr4 | IEP | | Rat | PMID:28108674|RGD:13463594 |
| Biological Process | GO:0014070 | response to organic cyclic compound | Cxcr4 | IMP | | Rat | PMID:21294880|RGD:6480473 |
| Biological Process | GO:1901327 | response to tacrolimus | Cxcr4 | IPI | CHEBI:61049 | Rat | PMID:24912495|RGD:151893463 |
| Biological Process | GO:1990478 | response to ultrasound | Cxcr4 | IEP | | Rat | PMID:25181476|RGD:11352664 |
| Biological Process | GO:0031290 | retinal ganglion cell axon guidance | cxcr4b | IGI | ZFIN:ZDB-GENO-070412-4,ZFIN:ZDB-MRPHLNO-050318-7 | Zfish | PMID:17267551 |
| Biological Process | GO:0061053 | somite development | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-071022-1 | Zfish | PMID:28701377 |
| Biological Process | GO:0061053 | somite development | cxcr4a | IGI | ZFIN:ZDB-GENE-020708-3 | Zfish | PMID:28701377 |
| Biological Process | GO:0061053 | somite development | cxcr4a | IGI | ZFIN:ZDB-GENE-030710-1 | Zfish | PMID:28701377 |
| Biological Process | GO:0002040 | sprouting angiogenesis | cxcr4a | IMP | ZFIN:ZDB-GENO-110513-3 | Zfish | PMID:21429983 |
| Biological Process | GO:0055002 | striated muscle cell development | cxcr4a | IMP | ZFIN:ZDB-MRPHLNO-071022-1 | Zfish | PMID:17517144 |
| Biological Process | GO:0042098 | T cell proliferation | Cxcr4 | IMP | MGI:MGI:1934442 | Mouse | PMID:12707343 |
| Biological Process | GO:0022029 | telencephalon cell migration | Cxcr4 | IMP | | Rat | PMID:16971524|RGD:2306584 |
| Biological Process | GO:0042246 | tissue regeneration | cxcr4b | IEP | | Zfish | PMID:16205622 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | cxcr4b | IMP | ZFIN:ZDB-GENO-071022-3 | Zfish | PMID:23382464 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | cxcr4b | IGI | ZFIN:ZDB-GENO-141204-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0001570 | vasculogenesis | cxcr4a | IMP | ZFIN:ZDB-GENO-091124-3 | Zfish | PMID:19797767 |
| Biological Process | GO:0003281 | ventricular septum development | Cxcr4 | IMP | | Mouse | PMID:25807483 |
 Analysis Tools
Analysis Tools