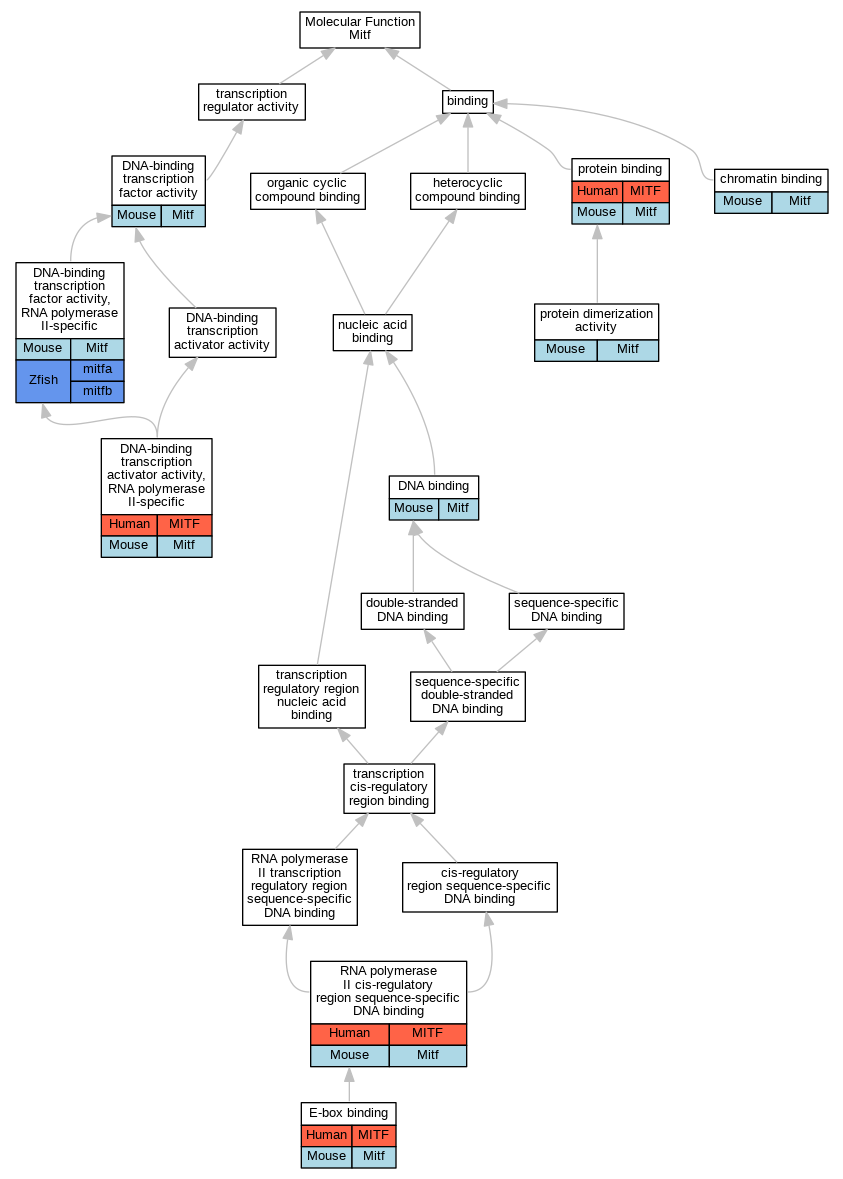
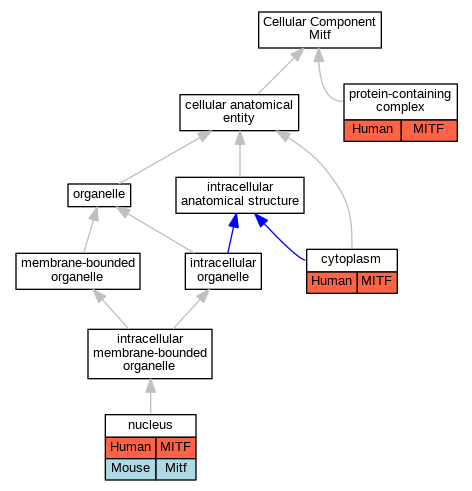
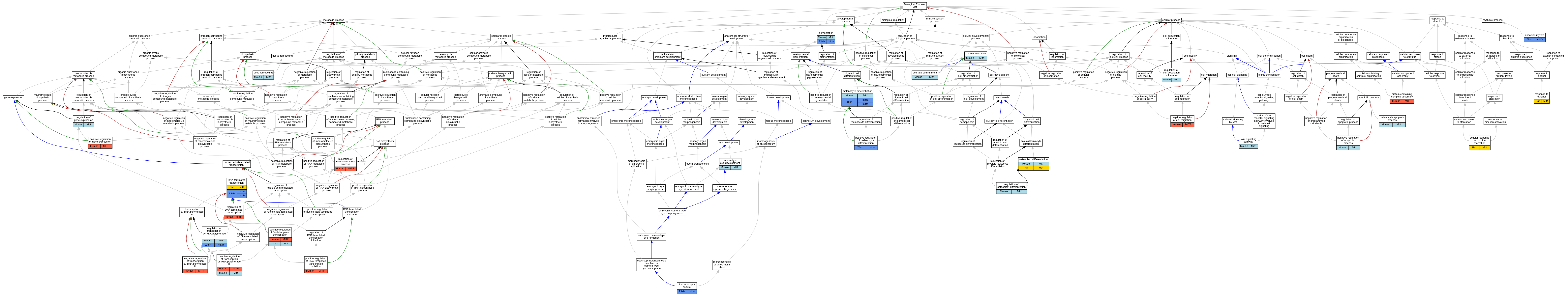
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003682 | chromatin binding | Mitf | IDA | | Mouse | PMID:15729346 |
| Molecular Function | GO:0003677 | DNA binding | Mitf | IDA | | Mouse | PMID:14575687 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | MITF | IDA | | Human | PMID:14737107 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | MITF | IMP | | Human | PMID:14737107 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | MITF | IDA | | Human | PMID:20530484 |
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | Mitf | IDA | | Mouse | MGI:MGI:3529668|PMID:15716956 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Mitf | IDA | | Mouse | PMID:12235125 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Mitf | IDA | | Mouse | PMID:14575687 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | Mitf | IMP | | Mouse | MGI:MGI:5461240|PMID:23207919 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | Mitf | IDA | | Mouse | PMID:15729346 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | mitfa | IDA | | Zfish | ZFIN:ZDB-PUB-010813-3|PMID:11543618 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | mitfb | IDA | | Zfish | PMID:11543618 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | mitfa | IDA | | Zfish | PMID:11543618 |
| Molecular Function | GO:0070888 | E-box binding | MITF | IDA | | Human | PMID:27889061 |
| Molecular Function | GO:0070888 | E-box binding | Mitf | IDA | | Mouse | MGI:MGI:5461240|PMID:23207919 |
| Molecular Function | GO:0005515 | protein binding | MITF | IPI | UniProtKB:P48436 | Human | PMID:20530484 |
| Molecular Function | GO:0005515 | protein binding | MITF | IPI | UniProtKB:Q92995 | Human | PMID:21811243 |
| Molecular Function | GO:0005515 | protein binding | MITF | IPI | UniProtKB:Q00610 | Human | PMID:35044719 |
| Molecular Function | GO:0005515 | protein binding | Mitf | IPI | PR:O54714 | Mouse | PMID:17442941 |
| Molecular Function | GO:0046983 | protein dimerization activity | Mitf | IPI | UniProtKB:Q08874 | Mouse | MGI:MGI:5461240|PMID:23207919 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | MITF | IDA | | Human | PMID:14737107 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Mitf | IDA | | Mouse | MGI:MGI:3529668|PMID:15716956 |
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | Mitf | IDA | | Mouse | PMID:23980096 |
| Cellular Component | GO:0005737 | cytoplasm | MITF | IDA | | Human | PMID:28842328 |
| Cellular Component | GO:0005634 | nucleus | MITF | IDA | | Human | PMID:27889061 |
| Cellular Component | GO:0005634 | nucleus | MITF | IDA | | Human | PMID:28842328 |
| Cellular Component | GO:0005634 | nucleus | Mitf | IDA | | Mouse | PMID:12235125 |
| Cellular Component | GO:0032991 | protein-containing complex | MITF | IDA | | Human | PMID:20530484 |
| Biological Process | GO:0046849 | bone remodeling | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0043010 | camera-type eye development | Mitf | IGI | MGI:MGI:88401 | Mouse | PMID:15576400 |
| Biological Process | GO:0030154 | cell differentiation | Mitf | IMP | MGI:MGI:1856094 | Mouse | PMID:9199364 |
| Biological Process | GO:0045165 | cell fate commitment | Mitf | IMP | MGI:MGI:1856085 | Mouse | PMID:15576400 |
| Biological Process | GO:0034224 | cellular response to zinc ion starvation | Mitf | IEP | | Rat | PMID:21893222|RGD:151667428 |
| Biological Process | GO:0007623 | circadian rhythm | mitfa | IDA | | Zfish | PMID:26278158 |
| Biological Process | GO:0061386 | closure of optic fissure | mitfa | IGI | ZFIN:ZDB-GENO-211027-12 | Zfish | PMID:32541011 |
| Biological Process | GO:0006351 | DNA-templated transcription | Mitf | IDA | | Rat | PMID:19524539|RGD:12793003 |
| Biological Process | GO:0006351 | DNA-templated transcription | mitfb | IDA | | Zfish | PMID:11543618|ZFIN:ZDB-PUB-010813-3 |
| Biological Process | GO:0006351 | DNA-templated transcription | mitfb | IDA | | Zfish | PMID:11543618 |
| Biological Process | GO:0006351 | DNA-templated transcription | mitfa | IDA | | Zfish | PMID:11543618 |
| Biological Process | GO:1902362 | melanocyte apoptotic process | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0030318 | melanocyte differentiation | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0030318 | melanocyte differentiation | Mitf | IGI | MGI:MGI:88138 | Mouse | PMID:12086670 |
| Biological Process | GO:0030318 | melanocyte differentiation | Mitf | IMP | MGI:MGI:1856088 | Mouse | PMID:2379821 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-110330-4 | Zfish | PMID:28249010 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IGI | ZFIN:ZDB-GENE-011212-6 | Zfish | PMID:28249010 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfb | IGI | ZFIN:ZDB-GENE-990910-11 | Zfish | PMID:11543618 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IGI | ZFIN:ZDB-MRPHLNO-060927-2,ZFIN:ZDB-MRPHLNO-101013-2 | Zfish | PMID:20862309 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-990423-18 | Zfish | PMID:10433906 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-070315-1 | Zfish | PMID:20862309 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-110330-3 | Zfish | PMID:21146516 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-100727-8 | Zfish | PMID:25535837 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | | Zfish | PMID:11543618 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-100727-8 | Zfish | PMID:21146516 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-MRPHLNO-050201-1 | Zfish | PMID:15300437 |
| Biological Process | GO:0030318 | melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-MRPHLNO-130429-1 | Zfish | PMID:23364329 |
| Biological Process | GO:0043066 | negative regulation of apoptotic process | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0030336 | negative regulation of cell migration | MITF | IMP | | Human | PMID:19188590 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | MITF | IMP | | Human | PMID:24769727 |
| Biological Process | GO:0030316 | osteoclast differentiation | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0030316 | osteoclast differentiation | Mitf | IEP | | Rat | PMID:22576626|RGD:151667429 |
| Biological Process | GO:0043473 | pigmentation | Mitf | IGI | MGI:MGI:88138 | Mouse | PMID:12086670 |
| Biological Process | GO:0043473 | pigmentation | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0043473 | pigmentation | Mitf | IMP | MGI:MGI:1856088 | Mouse | PMID:2379821 |
| Biological Process | GO:0043473 | pigmentation | mitfa | IMP | ZFIN:ZDB-GENO-100727-8 | Zfish | PMID:25214630 |
| Biological Process | GO:0043473 | pigmentation | mitfa | IGI | ZFIN:ZDB-GENE-011212-6 | Zfish | PMID:28249010 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | MITF | IDA | | Human | PMID:9647758 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Mitf | IDA | | Mouse | PMID:14575687 |
| Biological Process | GO:2000144 | positive regulation of DNA-templated transcription initiation | MITF | IDA | | Human | PMID:12204775 |
| Biological Process | GO:2000144 | positive regulation of DNA-templated transcription initiation | MITF | IDA | | Human | PMID:8995290 |
| Biological Process | GO:0010628 | positive regulation of gene expression | MITF | IDA | | Human | PMID:22234890 |
| Biological Process | GO:0010628 | positive regulation of gene expression | MITF | IMP | | Human | PMID:22234890 |
| Biological Process | GO:0010628 | positive regulation of gene expression | MITF | IMP | | Human | PMID:22523078 |
| Biological Process | GO:0045636 | positive regulation of melanocyte differentiation | mitfa | IMP | ZFIN:ZDB-GENO-100727-8 | Zfish | PMID:33439865 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | MITF | IMP | | Human | PMID:14737107 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | MITF | IDA | | Human | PMID:20530484 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | MITF | IDA | | Human | PMID:21209915 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mitf | IGI | UniProtKB:Q9R0G7 | Mouse | MGI:MGI:5574413|PMID:24769727 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mitf | IDA | | Mouse | MGI:MGI:5306313|PMID:15304486 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mitf | IDA | | Mouse | MGI:MGI:3529668|PMID:15716956 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mitf | IGI | MGI:MGI:98358 | Mouse | PMID:15729346 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Mitf | IDA | | Mouse | PMID:17442941 |
| Biological Process | GO:0065003 | protein-containing complex assembly | MITF | IDA | | Human | PMID:20530484 |
| Biological Process | GO:0042127 | regulation of cell population proliferation | Mitf | IDA | | Mouse | PMID:12235125 |
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | MITF | IMP | | Human | PMID:27889061 |
| Biological Process | GO:0010468 | regulation of gene expression | Mitf | IMP | MGI:MGI:1856095 | Mouse | PMID:12086670 |
| Biological Process | GO:0045670 | regulation of osteoclast differentiation | Mitf | IGI | MGI:MGI:98511 | Mouse | PMID:11930005 |
| Biological Process | GO:0045670 | regulation of osteoclast differentiation | Mitf | IMP | MGI:MGI:1856085 | Mouse | PMID:11930005 |
| Biological Process | GO:2001141 | regulation of RNA biosynthetic process | MITF | IDA | | Human | PMID:16411896 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Mitf | IMP | | Mouse | MGI:MGI:5461240|PMID:23207919 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Mitf | IDA | | Mouse | PMID:12235125 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | mitfb | IDA | | Zfish | PMID:11543618|ZFIN:ZDB-PUB-010813-3 |
| Biological Process | GO:0045471 | response to ethanol | Mitf | IEP | | Rat | PMID:22576626|RGD:151667429 |
| Biological Process | GO:0016055 | Wnt signaling pathway | Mitf | IDA | | Mouse | PMID:12235125 |
 Analysis Tools
Analysis Tools