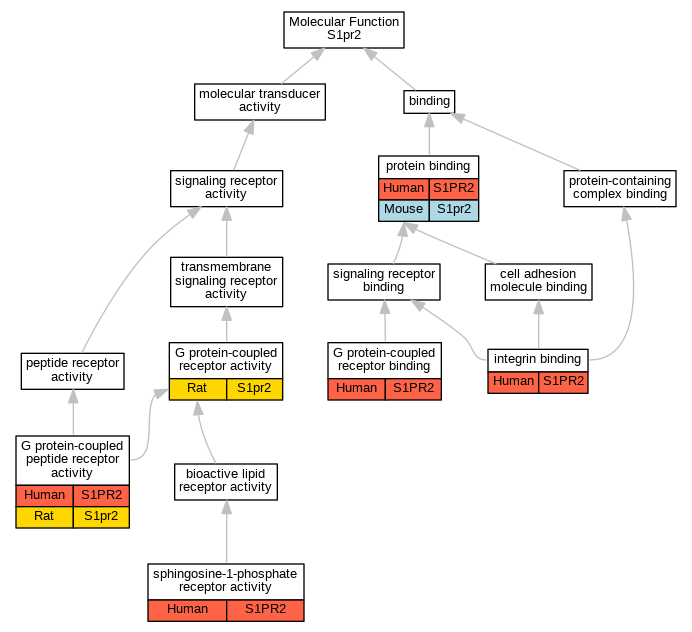
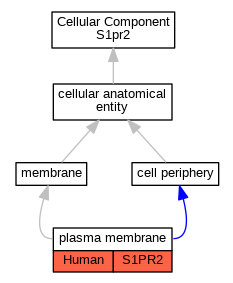
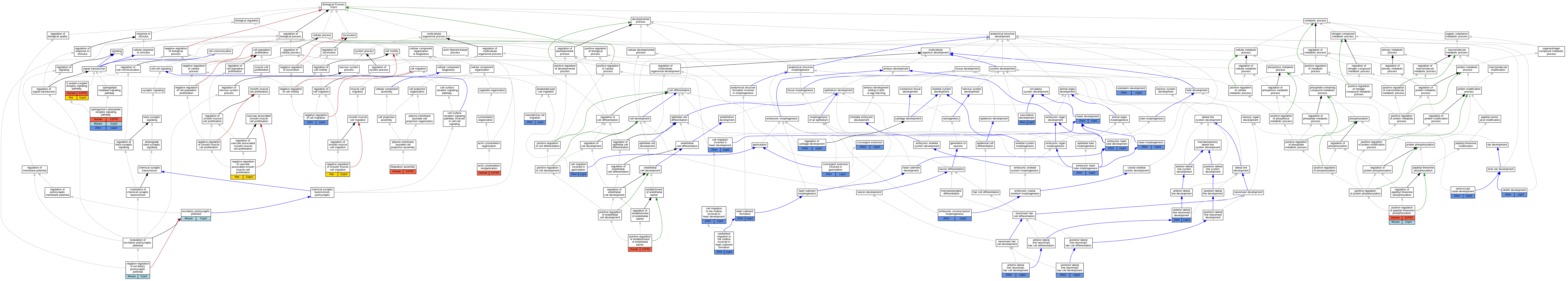
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0008528 | G protein-coupled peptide receptor activity | S1PR2 | IPI | UniProtKB:Q7Z5A7 | Human | PMID:29453251 |
| Molecular Function | GO:0008528 | G protein-coupled peptide receptor activity | S1pr2 | IDA | | Rat | PMID:29453251|RGD:42721986 |
| Molecular Function | GO:0004930 | G protein-coupled receptor activity | S1pr2 | IDA | | Rat | PMID:9409733|RGD:632521 |
| Molecular Function | GO:0001664 | G protein-coupled receptor binding | S1PR2 | IPI | UniProtKB:P21453 | Human | PMID:24851274 |
| Molecular Function | GO:0005178 | integrin binding | S1PR2 | IPI | UniProtKB:P16144 | Human | PMID:24851274 |
| Molecular Function | GO:0005515 | protein binding | S1PR2 | IPI | UniProtKB:Q9JK11-1 | Human | PMID:24453941 |
| Molecular Function | GO:0005515 | protein binding | S1PR2 | IPI | UniProtKB:P16144 | Human | PMID:24851274 |
| Molecular Function | GO:0005515 | protein binding | S1pr2 | IPI | UniProtKB:Q99P72|UniProtKB:Q9JK11-1 | Mouse | MGI:MGI:5563306|PMID:24453941 |
| Molecular Function | GO:0038036 | sphingosine-1-phosphate receptor activity | S1PR2 | IMP | | Human | PMID:23106337 |
| Molecular Function | GO:0038036 | sphingosine-1-phosphate receptor activity | S1PR2 | IDA | | Human | PMID:29453251 |
| Cellular Component | GO:0005886 | plasma membrane | S1PR2 | IDA | | Human | PMID:24851274 |
| Cellular Component | GO:0005886 | plasma membrane | S1PR2 | IDA | | Human | PMID:29453251 |
| Biological Process | GO:0031532 | actin cytoskeleton reorganization | S1PR2 | IMP | | Human | PMID:23106337 |
| Biological Process | GO:0048901 | anterior lateral line neuromast development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24176858 |
| Biological Process | GO:0035676 | anterior lateral line neuromast hair cell development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24176858 |
| Biological Process | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation | s1pr2 | IMP | ZFIN:ZDB-GENO-140411-5 | Zfish | PMID:23951404 |
| Biological Process | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation | s1pr2 | IGI | ZFIN:ZDB-GENO-140411-5,ZFIN:ZDB-MRPHLNO-050620-2 | Zfish | PMID:23951404 |
| Biological Process | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation | s1pr2 | IGI | ZFIN:ZDB-GENO-130530-5,ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:23951404 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:18701549 |
| Biological Process | GO:0042074 | cell migration involved in gastrulation | s1pr2 | IGI | ZFIN:ZDB-MRPHLNO-070911-3,ZFIN:ZDB-MRPHLNO-081222-1 | Zfish | PMID:18701549 |
| Biological Process | GO:0060973 | cell migration involved in heart development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:23318642 |
| Biological Process | GO:0060973 | cell migration involved in heart development | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:23318642 |
| Biological Process | GO:0003318 | cell migration to the midline involved in heart development | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:25997406 |
| Biological Process | GO:0060026 | convergent extension | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:23318642 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | s1pr2 | IDA | | Zfish | PMID:19062281 |
| Biological Process | GO:0035050 | embryonic heart tube development | s1pr2 | IMP | ZFIN:ZDB-TALEN-131219-2 | Zfish | PMID:23616919 |
| Biological Process | GO:0035050 | embryonic heart tube development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:23616919 |
| Biological Process | GO:0003143 | embryonic heart tube morphogenesis | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24451492 |
| Biological Process | GO:0003143 | embryonic heart tube morphogenesis | s1pr2 | IGI | ZFIN:ZDB-GENE-040426-1156,ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24451492 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | s1pr2 | IGI | ZFIN:ZDB-GENO-140411-5,ZFIN:ZDB-MRPHLNO-050620-2 | Zfish | PMID:23951404 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | s1pr2 | IGI | ZFIN:ZDB-GENO-130530-5,ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:23951404 |
| Biological Process | GO:0007492 | endoderm development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:23318642 |
| Biological Process | GO:0007492 | endoderm development | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:23318642 |
| Biological Process | GO:0060079 | excitatory postsynaptic potential | S1pr2 | IMP | MGI:MGI:2180817 | Mouse | PMID:11553273 |
| Biological Process | GO:0046847 | filopodium assembly | S1PR2 | IMP | | Human | PMID:23106337 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | S1PR2 | IDA | | Human | PMID:29453251 |
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | S1pr2 | IDA | | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:0007507 | heart development | s1pr2 | IMP | ZFIN:ZDB-GENO-980202-969 | Zfish | PMID:17537802 |
| Biological Process | GO:0003007 | heart morphogenesis | s1pr2 | IMP | ZFIN:ZDB-GENO-980202-969 | Zfish | PMID:9007249 |
| Biological Process | GO:0003007 | heart morphogenesis | s1pr2 | IGI | ZFIN:ZDB-GENO-070911-2,ZFIN:ZDB-MRPHLNO-070911-2 | Zfish | PMID:17230219 |
| Biological Process | GO:0003007 | heart morphogenesis | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:9007249 |
| Biological Process | GO:0003007 | heart morphogenesis | s1pr2 | IMP | ZFIN:ZDB-GENO-070911-2 | Zfish | PMID:17230219 |
| Biological Process | GO:0003315 | heart rudiment formation | s1pr2 | IGI | ZFIN:ZDB-GENO-010411-9,ZFIN:ZDB-MRPHLNO-090730-1 | Zfish | PMID:25555130 |
| Biological Process | GO:0003315 | heart rudiment formation | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:25555130 |
| Biological Process | GO:0008078 | mesodermal cell migration | s1pr2 | IGI | ZFIN:ZDB-GENO-070911-1,ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:18701549 |
| Biological Process | GO:0030336 | negative regulation of cell migration | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:18701549 |
| Biological Process | GO:0030336 | negative regulation of cell migration | s1pr2 | IGI | ZFIN:ZDB-MRPHLNO-070911-3,ZFIN:ZDB-MRPHLNO-081222-1 | Zfish | PMID:18701549 |
| Biological Process | GO:0090394 | negative regulation of excitatory postsynaptic potential | S1pr2 | IMP | MGI:MGI:2180817 | Mouse | PMID:11553273 |
| Biological Process | GO:0014912 | negative regulation of smooth muscle cell migration | S1pr2 | IGI | RGD:1562115 | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:1904706 | negative regulation of vascular associated smooth muscle cell proliferation | S1pr2 | IGI | RGD:1562115 | Rat | PMID:29453251|RGD:42721986 |
| Biological Process | GO:0048840 | otolith development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24176858 |
| Biological Process | GO:1903142 | positive regulation of establishment of endothelial barrier | S1PR2 | IMP | | Human | PMID:24851274 |
| Biological Process | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | S1PR2 | IMP | | Human | PMID:23106337 |
| Biological Process | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation | S1pr2 | IMP | | Mouse | MGI:MGI:5470025|PMID:23106337 |
| Biological Process | GO:0035677 | posterior lateral line neuromast hair cell development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24176858 |
| Biological Process | GO:0061035 | regulation of cartilage development | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:22185793 |
| Biological Process | GO:0060872 | semicircular canal development | s1pr2 | IMP | ZFIN:ZDB-MRPHLNO-070911-3 | Zfish | PMID:24176858 |
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | S1PR2 | IMP | | Human | PMID:23106337 |
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | S1pr2 | IMP | | Mouse | MGI:MGI:5470025|PMID:23106337 |
| Biological Process | GO:0003376 | sphingosine-1-phosphate receptor signaling pathway | s1pr2 | IMP | ZFIN:ZDB-GENO-010411-9 | Zfish | PMID:22185793 |
| Biological Process | GO:0001944 | vasculature development | s1pr2 | IGI | ZFIN:ZDB-MRPHLNO-070911-3,ZFIN:ZDB-MRPHLNO-121026-2 | Zfish | PMID:23229546 |
 Analysis Tools
Analysis Tools