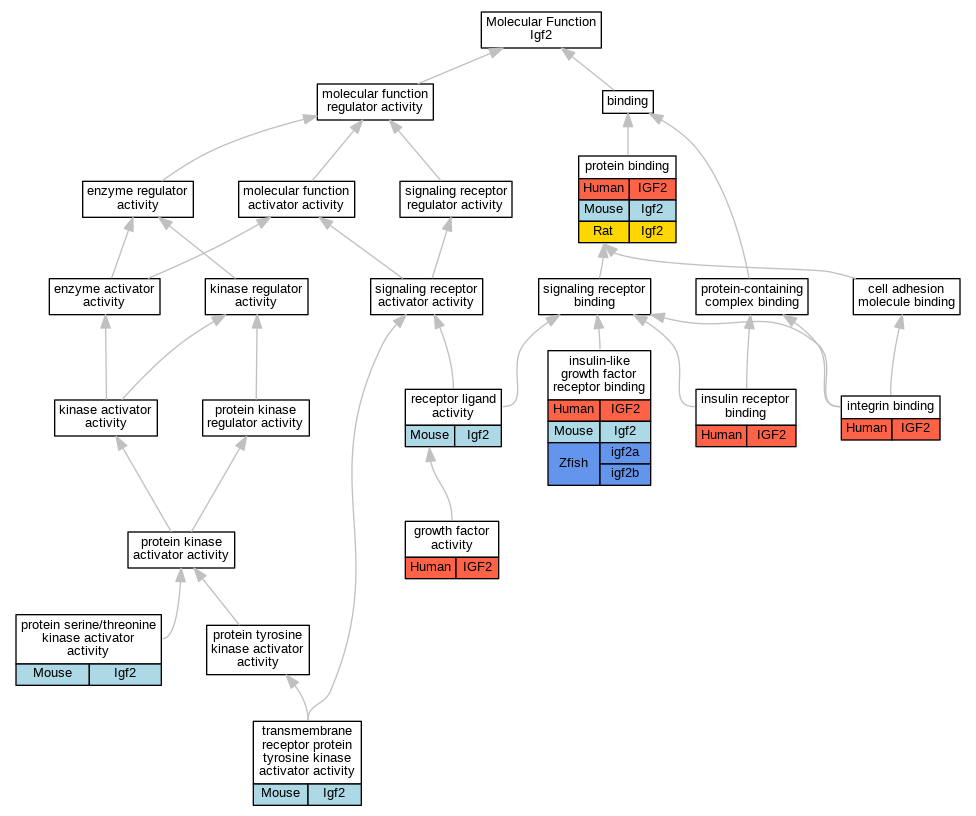
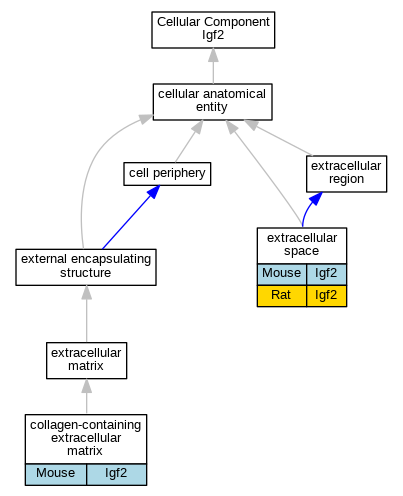
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0008083 | growth factor activity | IGF2 | IDA | | Human | PMID:11500939 |
| Molecular Function | GO:0005158 | insulin receptor binding | IGF2 | IPI | UniProtKB:P06213 | Human | PMID:12138094 |
| Molecular Function | GO:0005159 | insulin-like growth factor receptor binding | IGF2 | IMP | | Human | PMID:2967174 |
| Molecular Function | GO:0005159 | insulin-like growth factor receptor binding | Igf2 | IPI | PR:Q07113 | Mouse | PMID:14566968 |
| Molecular Function | GO:0005159 | insulin-like growth factor receptor binding | igf2b | IDA | | Zfish | PMID:19759899 |
| Molecular Function | GO:0005159 | insulin-like growth factor receptor binding | igf2a | IDA | | Zfish | PMID:19759899 |
| Molecular Function | GO:0005178 | integrin binding | IGF2 | IDA | | Human | PMID:28873464 |
| Molecular Function | GO:0005515 | protein binding | IGF2 | IPI | UniProtKB:Q9PTH3 | Human | PMID:10611375 |
| Molecular Function | GO:0005515 | protein binding | IGF2 | IPI | UniProtKB:P11717 | Human | PMID:18046459 |
| Molecular Function | GO:0005515 | protein binding | IGF2 | IPI | UniProtKB:Q93062 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | Igf2 | IPI | PR:Q69ZQ1 | Mouse | PMID:19706595 |
| Molecular Function | GO:0005515 | protein binding | Igf2 | IPI | PR:Q07079 | Mouse | PMID:14566968 |
| Molecular Function | GO:0005515 | protein binding | Igf2 | IPI | UniProtKB:P12843 | Rat | PMID:2538475|RGD:729148 |
| Molecular Function | GO:0005515 | protein binding | Igf2 | IPI | UniProtKB:P12843 | Rat | PMID:2974285|RGD:8553840 |
| Molecular Function | GO:0043539 | protein serine/threonine kinase activator activity | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Molecular Function | GO:0048018 | receptor ligand activity | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Molecular Function | GO:0048018 | receptor ligand activity | Igf2 | IGI | MGI:MGI:96433 | Mouse | PMID:9281335 |
| Molecular Function | GO:0048018 | receptor ligand activity | Igf2 | IDA | | Mouse | PMID:29348399 |
| Molecular Function | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity | Igf2 | IGI | MGI:MGI:96433 | Mouse | PMID:9281335 |
| Molecular Function | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity | Igf2 | IDA | | Mouse | PMID:29348399 |
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | Igf2 | HDA | | Mouse | MGI:MGI:6201853|PMID:28071719 |
| Cellular Component | GO:0005615 | extracellular space | Igf2 | IDA | | Mouse | PMID:18762576 |
| Cellular Component | GO:0005615 | extracellular space | Igf2 | IDA | | Rat | PMID:6390212|RGD:1624322 |
| Biological Process | GO:0009887 | animal organ morphogenesis | Igf2 | IMP | MGI:MGI:2677156 | Mouse | PMID:12101187 |
| Biological Process | GO:0071260 | cellular response to mechanical stimulus | Igf2 | IEP | | Rat | PMID:20695079|RGD:5509947 |
| Biological Process | GO:0001892 | embryonic placenta development | Igf2 | IMP | | Mouse | MGI:MGI:2181513|PMID:12087403 |
| Biological Process | GO:0060669 | embryonic placenta morphogenesis | Igf2 | IMP | | Mouse | MGI:MGI:5908476|PMID:28522536 |
| Biological Process | GO:0031017 | exocrine pancreas development | Igf2 | IMP | MGI:MGI:2677156 | Mouse | PMID:12101187 |
| Biological Process | GO:0007565 | female pregnancy | Igf2 | IEP | | Rat | PMID:19776249|RGD:5509949 |
| Biological Process | GO:0031101 | fin regeneration | igf2b | IMP | ZFIN:ZDB-MRPHLNO-100601-1 | Zfish | PMID:20179093 |
| Biological Process | GO:0030900 | forebrain development | igf2b | IMP | ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0001701 | in utero embryonic development | Igf2 | IMP | | Mouse | MGI:MGI:5908476|PMID:28522536 |
| Biological Process | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0048009 | insulin-like growth factor receptor signaling pathway | Igf2 | IGI | MGI:MGI:96433 | Mouse | PMID:9281335 |
| Biological Process | GO:0048009 | insulin-like growth factor receptor signaling pathway | Igf2 | IDA | | Mouse | PMID:29348399 |
| Biological Process | GO:0007613 | memory | Igf2 | IMP | | Rat | PMID:21270887|RGD:5509946 |
| Biological Process | GO:0051148 | negative regulation of muscle cell differentiation | Igf2 | IMP | | Mouse | MGI:MGI:6467708|PMID:32557799 |
| Biological Process | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity | Igf2 | IDA | | Rat | PMID:9021955|RGD:9685777 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | Igf2 | IMP | | Mouse | MGI:MGI:5908341|PMID:16901893 |
| Biological Process | GO:0030903 | notochord development | igf2b | IMP | ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0030903 | notochord development | igf2b | IGI | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2,ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0030903 | notochord development | igf2a | IGI | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2,ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0030903 | notochord development | igf2a | IMP | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2 | Zfish | PMID:19443571 |
| Biological Process | GO:0001649 | osteoblast differentiation | Igf2 | IEP | | Mouse | MGI:MGI:3828402|PMID:16311053 |
| Biological Process | GO:0042104 | positive regulation of activated T cell proliferation | IGF2 | IDA | | Human | PMID:15694994 |
| Biological Process | GO:0045766 | positive regulation of angiogenesis | Igf2 | IGI | RNAcentral:URS00004DE9D7_10116 | Rat | PMID:28735866|RGD:13506264 |
| Biological Process | GO:0043536 | positive regulation of blood vessel endothelial cell migration | Igf2 | IGI | RNAcentral:URS00004DE9D7_10116 | Rat | PMID:28735866|RGD:13506264 |
| Biological Process | GO:0043085 | positive regulation of catalytic activity | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IGF2 | IDA | | Human | PMID:28873464 |
| Biological Process | GO:2000467 | positive regulation of glycogen (starch) synthase activity | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0045725 | positive regulation of glycogen biosynthetic process | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0046628 | positive regulation of insulin receptor signaling pathway | IGF2 | IDA | | Human | PMID:11500939 |
| Biological Process | GO:0046628 | positive regulation of insulin receptor signaling pathway | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IGF2 | IDA | | Human | PMID:11500939 |
| Biological Process | GO:0045840 | positive regulation of mitotic nuclear division | IGF2 | IDA | | Human | PMID:11500939 |
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | Igf2 | IMP | | Mouse | PMID:9203585 |
| Biological Process | GO:0046622 | positive regulation of organ growth | Igf2 | IMP | | Mouse | MGI:MGI:6120647|PMID:29440408 |
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0051897 | positive regulation of protein kinase B signaling | IGF2 | IDA | | Human | PMID:11500939 |
| Biological Process | GO:0001934 | positive regulation of protein phosphorylation | Igf2 | IMP | | Mouse | MGI:MGI:4438697|PMID:20032056 |
| Biological Process | GO:0048633 | positive regulation of skeletal muscle tissue growth | Igf2 | IMP | | Mouse | MGI:MGI:6120647|PMID:29440408 |
| Biological Process | GO:0090031 | positive regulation of steroid hormone biosynthetic process | Igf2 | IDA | | Rat | PMID:17023532|RGD:1600072 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Igf2 | IGI | MGI:MGI:87986 | Mouse | PMID:18762576 |
| Biological Process | GO:1905564 | positive regulation of vascular endothelial cell proliferation | Igf2 | IGI | RNAcentral:URS00004DE9D7_10116 | Rat | PMID:28735866|RGD:13506264 |
| Biological Process | GO:0048793 | pronephros development | igf2b | IMP | ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0051147 | regulation of muscle cell differentiation | Igf2 | IMP | | Mouse | MGI:MGI:5908341|PMID:16901893 |
| Biological Process | GO:0032355 | response to estradiol | Igf2 | IEP | | Rat | PMID:18946176|RGD:2311518 |
| Biological Process | GO:0045471 | response to ethanol | Igf2 | IEP | | Rat | PMID:18505416|RGD:5510002 |
| Biological Process | GO:0035094 | response to nicotine | Igf2 | IDA | | Rat | PMID:19494366|RGD:5509950 |
| Biological Process | GO:0035094 | response to nicotine | Igf2 | IEP | | Rat | PMID:19494366|RGD:5509950 |
| Biological Process | GO:0031667 | response to nutrient levels | Igf2 | IEP | | Rat | PMID:17626604|RGD:2311522 |
| Biological Process | GO:0014070 | response to organic cyclic compound | Igf2 | IDA | | Rat | PMID:17407073|RGD:2311525 |
| Biological Process | GO:0009314 | response to radiation | Igf2 | IEP | | Rat | PMID:17630476|RGD:2311521 |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Igf2 | IDA | | Rat | PMID:18778817|RGD:5509951 |
| Biological Process | GO:0001756 | somitogenesis | igf2b | IMP | ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0001756 | somitogenesis | igf2b | IGI | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2,ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0001756 | somitogenesis | igf2a | IMP | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2 | Zfish | PMID:19443571 |
| Biological Process | GO:0001756 | somitogenesis | igf2a | IGI | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2,ZFIN:ZDB-MRPHLNO-090929-3,ZFIN:ZDB-MRPHLNO-090929-4 | Zfish | PMID:19443571 |
| Biological Process | GO:0060720 | spongiotrophoblast cell proliferation | Igf2 | IMP | | Mouse | MGI:MGI:5424276|PMID:22562201 |
| Biological Process | GO:0002040 | sprouting angiogenesis | igf2b | IMP | ZFIN:ZDB-MRPHLNO-090929-4,ZFIN:ZDB-MRPHLNO-100527-1 | Zfish | PMID:29951828 |
| Biological Process | GO:0002040 | sprouting angiogenesis | igf2a | IMP | ZFIN:ZDB-MRPHLNO-090929-1,ZFIN:ZDB-MRPHLNO-090929-2 | Zfish | PMID:29951828 |
| Biological Process | GO:0051146 | striated muscle cell differentiation | Igf2 | IGI | MGI:MGI:96440 | Mouse | PMID:18762576 |
| Biological Process | GO:0042060 | wound healing | Igf2 | IEP | | Rat | PMID:7770004|RGD:5509999 |
 Analysis Tools
Analysis Tools