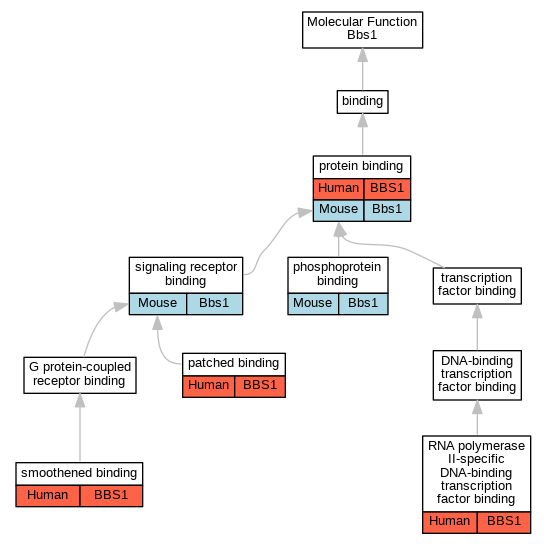
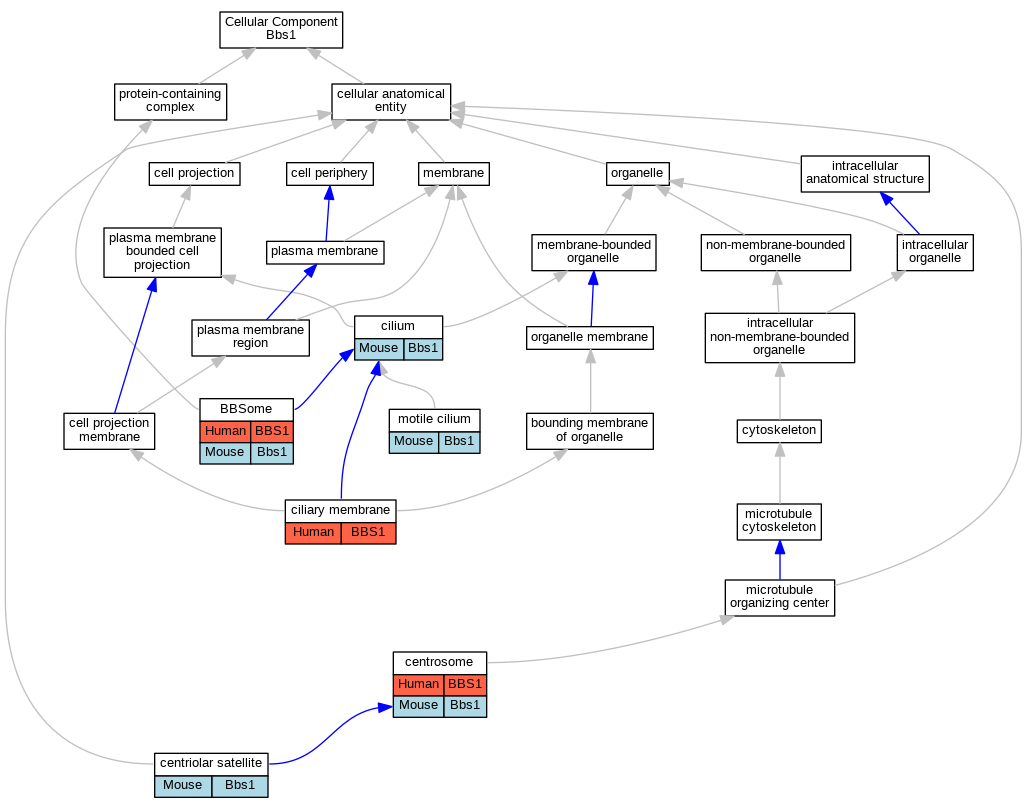
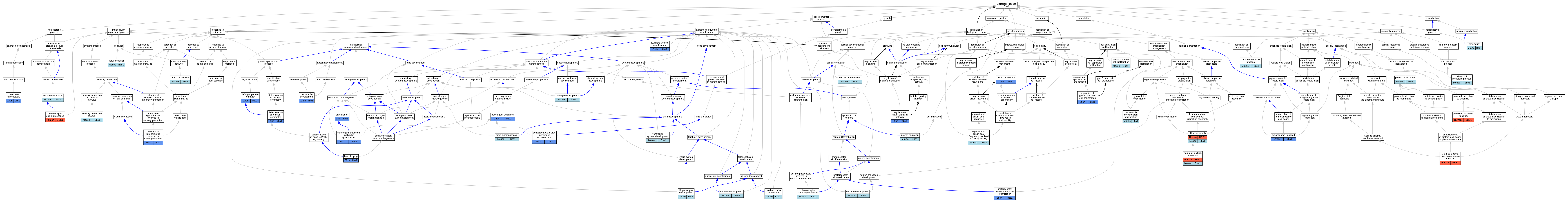
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005113 | patched binding | BBS1 | IPI | UniProtKB:Q13635 | Human | PMID:22228099 |
| Molecular Function | GO:0051219 | phosphoprotein binding | Bbs1 | IPI | PR:Q811T9-1 | Mouse | PMID:21471969 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9BUN5 | Human | PMID:16327777 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q15154 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q3SYG4 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96QF0-1 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96QF0-2 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96RK4 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9BXC9 | Human | PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:P05062 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:P68104 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q15154 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q99497 | Human | PMID:18000879 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9NRI5 | Human | PMID:18762586 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96RK4 | Human | PMID:19081074 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:P48356-1 | Human | PMID:19150989 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q3SYG4 | Human | PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9H0F7 | Human | PMID:20603001 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9H0F7 | Human | PMID:22139371 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q3SYG4 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96RK4 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9BXC9 | Human | PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q8TAM2 | Human | PMID:24939912 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9H0F7-1 | Human | PMID:25402481 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q15051 | Human | PMID:25552655 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q3SYG4 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q8IWZ6 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q96RK4 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q9BXC9 | Human | PMID:27173435 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q3SYG4 | Human | PMID:29039417 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q14203-5 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | BBS1 | IPI | UniProtKB:Q99497 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | UniProtKB:Q8C1Z7 | Mouse | MGI:MGI:4420847|PMID:20080638 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | UniProtKB:Q8C1Z7 | Mouse | MGI:MGI:5467205|PMID:22500027 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | UniProtKB:Q8C1Z7 | Mouse | MGI:MGI:5306530|PMID:22139371 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | UniProtKB:Q99497 | Mouse | MGI:MGI:5574296|PMID:21097510 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | UniProtKB:Q8C1Z7 | Mouse | MGI:MGI:5926591|PMID:17574030 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | PR:Q9R0L6 | Mouse | PMID:27979967 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | PR:Q8BMD2 | Mouse | PMID:27979967 |
| Molecular Function | GO:0005515 | protein binding | Bbs1 | IPI | PR:Q8C1Z7 | Mouse | PMID:27979967 |
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | BBS1 | IPI | UniProtKB:Q99496 | Human | PMID:22302990 |
| Molecular Function | GO:0005102 | signaling receptor binding | Bbs1 | IPI | PR:P48356 | Mouse | PMID:19150989 |
| Molecular Function | GO:0005119 | smoothened binding | BBS1 | IPI | UniProtKB:Q99835 | Human | PMID:22228099 |
| Cellular Component | GO:0034464 | BBSome | BBS1 | IPI | | Human | PMID:19081074 |
| Cellular Component | GO:0034464 | BBSome | BBS1 | IDA | | Human | PMID:17574030 |
| Cellular Component | GO:0034464 | BBSome | BBS1 | IDA | | Human | PMID:20080638 |
| Cellular Component | GO:0034464 | BBSome | BBS1 | IDA | | Human | PMID:24550735 |
| Cellular Component | GO:0034464 | BBSome | Bbs1 | IDA | | Mouse | PMID:20080638 |
| Cellular Component | GO:0034464 | BBSome | Bbs1 | IDA | | Mouse | PMID:22500027 |
| Cellular Component | GO:0034464 | BBSome | Bbs1 | IDA | | Mouse | PMID:22072986 |
| Cellular Component | GO:0034451 | centriolar satellite | Bbs1 | IDA | | Mouse | PMID:27979967 |
| Cellular Component | GO:0005813 | centrosome | BBS1 | IDA | | Human | PMID:18762586 |
| Cellular Component | GO:0005813 | centrosome | Bbs1 | IDA | | Mouse | PMID:21471969 |
| Cellular Component | GO:0060170 | ciliary membrane | BBS1 | IDA | | Human | PMID:19081074 |
| Cellular Component | GO:0005929 | cilium | Bbs1 | IDA | | Mouse | PMID:27979967 |
| Cellular Component | GO:0031514 | motile cilium | Bbs1 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Biological Process | GO:0030534 | adult behavior | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0048854 | brain morphogenesis | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0051216 | cartilage development | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:19195025 |
| Biological Process | GO:0044255 | cellular lipid metabolic process | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0021987 | cerebral cortex development | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0042632 | cholesterol homeostasis | bbs1 | IMP | ZFIN:ZDB-GENO-221107-2 | Zfish | PMID:35277505 |
| Biological Process | GO:0042632 | cholesterol homeostasis | bbs1 | IMP | ZFIN:ZDB-GENO-221107-1 | Zfish | PMID:35277505 |
| Biological Process | GO:0060271 | cilium assembly | BBS1 | IMP | | Human | PMID:17574030 |
| Biological Process | GO:0060271 | cilium assembly | Bbs1 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Biological Process | GO:0060271 | cilium assembly | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0060271 | cilium assembly | Bbs1 | IMP | | Mouse | PMID:27979967 |
| Biological Process | GO:0003341 | cilium movement | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:24069149 |
| Biological Process | GO:0060026 | convergent extension | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-140729-1 | Zfish | PMID:24691443 |
| Biological Process | GO:0060026 | convergent extension | bbs1 | IGI | ZFIN:ZDB-GENO-080222-3,ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:17906624 |
| Biological Process | GO:0060026 | convergent extension | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:17906624 |
| Biological Process | GO:0060026 | convergent extension | bbs1 | IGI | ZFIN:ZDB-GENO-080222-4,ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:17906624 |
| Biological Process | GO:0060028 | convergent extension involved in axis elongation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:24069149 |
| Biological Process | GO:0060027 | convergent extension involved in gastrulation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:16582908 |
| Biological Process | GO:0016358 | dendrite development | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:15322545 |
| Biological Process | GO:0050908 | detection of light stimulus involved in visual perception | bbs1 | IMP | ZFIN:ZDB-GENO-221107-2 | Zfish | PMID:35277505 |
| Biological Process | GO:0050908 | detection of light stimulus involved in visual perception | bbs1 | IMP | ZFIN:ZDB-GENO-221107-3 | Zfish | PMID:35277505 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-081027-1,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060320-6,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060519-1,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0007368 | determination of left/right symmetry | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-5,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0045444 | fat cell differentiation | Bbs1 | IEP | | Mouse | MGI:MGI:3794016|PMID:17379567 |
| Biological Process | GO:0009566 | fertilization | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0007369 | gastrulation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:20498079 |
| Biological Process | GO:0007369 | gastrulation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-140729-1 | Zfish | PMID:20498079 |
| Biological Process | GO:0043001 | Golgi to plasma membrane protein transport | BBS1 | IMP | | Human | PMID:19150989 |
| Biological Process | GO:0001947 | heart looping | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:24069149 |
| Biological Process | GO:0021766 | hippocampus development | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0042445 | hormone metabolic process | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060519-1,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-081027-1,ZFIN:ZDB-MRPHLNO-081223-2 | Zfish | PMID:19081074 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-081027-1,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060320-6,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0070121 | Kupffer's vesicle development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-5,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0060972 | left/right pattern formation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:24069149 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-081027-1,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-2 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060320-6,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-5,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060519-1,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0032402 | melanosome transport | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060209-3,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0000226 | microtubule cytoskeleton organization | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:15322545 |
| Biological Process | GO:0061351 | neural precursor cell proliferation | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:23160237 |
| Biological Process | GO:0001764 | neuron migration | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:21471969 |
| Biological Process | GO:1905515 | non-motile cilium assembly | BBS1 | IMP | | Human | PMID:17980398 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Bbs1 | IGI | MGI:MGI:2135272 | Mouse | PMID:16170314 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:19195025 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:16170314 |
| Biological Process | GO:1905515 | non-motile cilium assembly | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:15322545 |
| Biological Process | GO:0042048 | olfactory behavior | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0033339 | pectoral fin development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-060320-6,ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0033339 | pectoral fin development | bbs1 | IGI | ZFIN:ZDB-MRPHLNO-081027-1,ZFIN:ZDB-MRPHLNO-081027-3 | Zfish | PMID:18381349 |
| Biological Process | GO:0033339 | pectoral fin development | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-081027-1 | Zfish | PMID:18381349 |
| Biological Process | GO:0045494 | photoreceptor cell maintenance | BBS1 | IMP | | Human | PMID:17980398 |
| Biological Process | GO:0008594 | photoreceptor cell morphogenesis | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0035845 | photoreceptor cell outer segment organization | bbs1 | IMP | ZFIN:ZDB-GENO-221107-2 | Zfish | PMID:35277505 |
| Biological Process | GO:0035845 | photoreceptor cell outer segment organization | bbs1 | IMP | ZFIN:ZDB-GENO-221107-3 | Zfish | PMID:35277505 |
| Biological Process | GO:0035845 | photoreceptor cell outer segment organization | bbs1 | IMP | ZFIN:ZDB-GENO-221107-1 | Zfish | PMID:35277505 |
| Biological Process | GO:0008104 | protein localization | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:22228099 |
| Biological Process | GO:0061512 | protein localization to cilium | BBS1 | IMP | | Human | PMID:23943788 |
| Biological Process | GO:0060296 | regulation of cilium beat frequency involved in ciliary motility | Bbs1 | IMP | | Mouse | MGI:MGI:3776933|PMID:18299575 |
| Biological Process | GO:0008593 | regulation of Notch signaling pathway | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:24681783 |
| Biological Process | GO:0061469 | regulation of type B pancreatic cell proliferation | bbs1 | IMP | ZFIN:ZDB-MRPHLNO-060208-2 | Zfish | PMID:26494903 |
| Biological Process | GO:0001895 | retina homeostasis | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:15322545 |
| Biological Process | GO:0007608 | sensory perception of smell | Bbs1 | IMP | MGI:MGI:3055491 | Mouse | PMID:15322545 |
| Biological Process | GO:0021756 | striatum development | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:18032602 |
| Biological Process | GO:0021591 | ventricular system development | Bbs1 | IMP | MGI:MGI:3767672 | Mouse | PMID:23160237 |
| Biological Process | GO:0021591 | ventricular system development | Bbs1 | IMP | MGI:MGI:4837746,MGI:MGI:5474481 | Mouse | PMID:23160237 |
 Analysis Tools
Analysis Tools