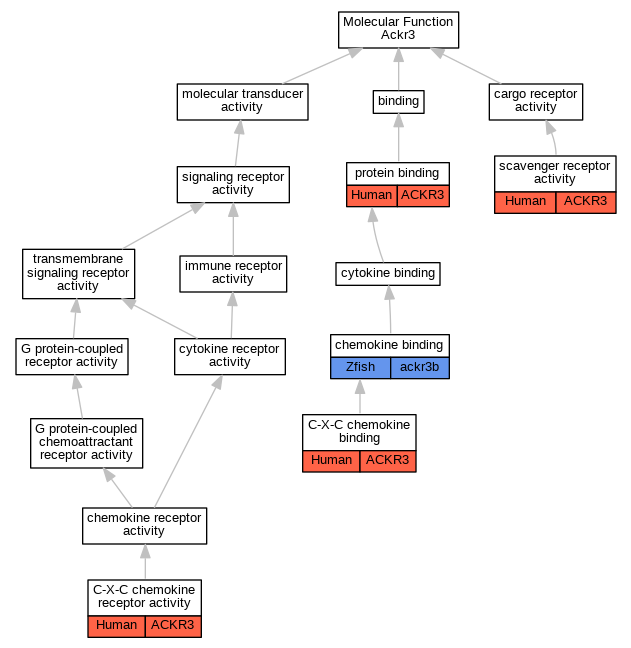
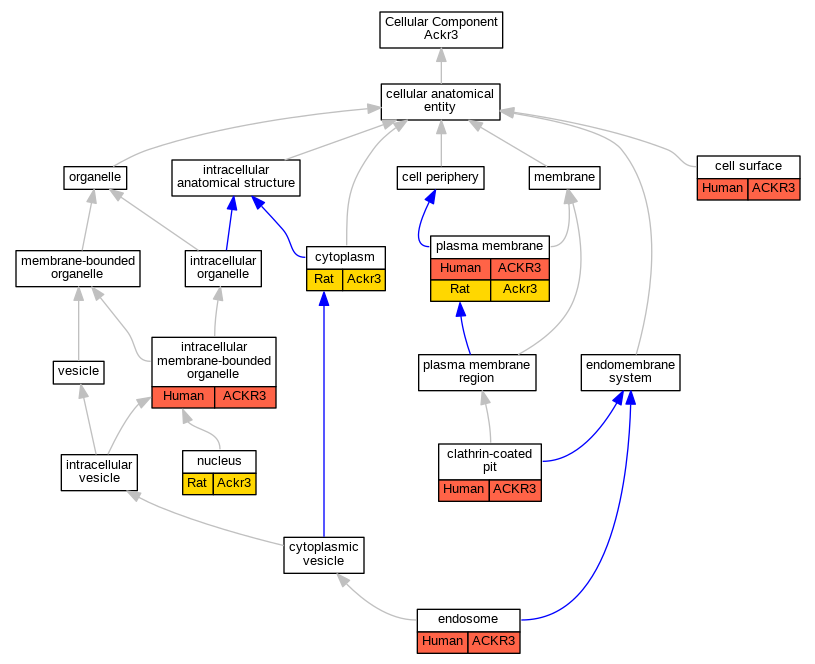
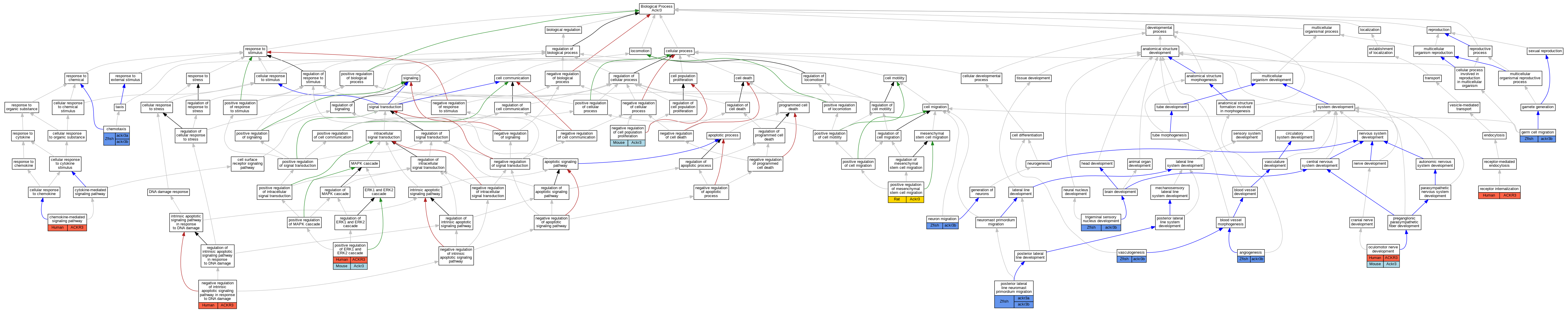
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0019958 | C-X-C chemokine binding | ACKR3 | IMP | | Human | PMID:22300987 |
| Molecular Function | GO:0019958 | C-X-C chemokine binding | ACKR3 | IMP | | Human | PMID:31211835 |
| Molecular Function | GO:0016494 | C-X-C chemokine receptor activity | ACKR3 | IMP | | Human | PMID:20388803 |
| Molecular Function | GO:0019956 | chemokine binding | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-080725-5 | Zfish | PMID:18267076 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:19380869 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:21730065 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P32121 | Human | PMID:22300987 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P32121 | Human | PMID:22457824 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P49407 | Human | PMID:22457824 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:25775528 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:27331810 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:28862946 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P37288 | Human | PMID:29386406 |
| Molecular Function | GO:0005515 | protein binding | ACKR3 | IPI | UniProtKB:P61073 | Human | PMID:29386406 |
| Molecular Function | GO:0005044 | scavenger receptor activity | ACKR3 | IMP | | Human | PMID:22300987 |
| Cellular Component | GO:0009986 | cell surface | ACKR3 | IDA | | Human | PMID:22457824 |
| Cellular Component | GO:0005905 | clathrin-coated pit | ACKR3 | IDA | | Human | PMID:22457824 |
| Cellular Component | GO:0005737 | cytoplasm | Ackr3 | IDA | | Rat | PMID:24924806|RGD:11352662 |
| Cellular Component | GO:0005768 | endosome | ACKR3 | IDA | | Human | PMID:22300987 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ACKR3 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | Ackr3 | IDA | | Rat | PMID:24924806|RGD:11352662 |
| Cellular Component | GO:0005886 | plasma membrane | ACKR3 | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005886 | plasma membrane | ACKR3 | IDA | | Human | PMID:22300987 |
| Cellular Component | GO:0005886 | plasma membrane | ACKR3 | IDA | | Human | PMID:31211835 |
| Cellular Component | GO:0005886 | plasma membrane | Ackr3 | IDA | | Rat | PMID:24924806|RGD:11352662 |
| Biological Process | GO:0001525 | angiogenesis | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-071105-1,ZFIN:ZDB-MRPHLNO-071105-2 | Zfish | PMID:17898181 |
| Biological Process | GO:0070098 | chemokine-mediated signaling pathway | ACKR3 | IMP | | Human | PMID:20388803 |
| Biological Process | GO:0006935 | chemotaxis | ackr3b | IGI | ZFIN:ZDB-GENO-100507-5,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | ackr3b | IMP | ZFIN:ZDB-GENO-100507-5 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | ackr3b | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | ackr3a | IGI | ZFIN:ZDB-GENO-100507-5,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | ackr3a | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0006935 | chemotaxis | ackr3a | IMP | ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0008354 | germ cell migration | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-080725-5 | Zfish | PMID:18267076 |
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | Ackr3 | IGI | MGI:MGI:103556,MGI:MGI:1341264 | Mouse | PMID:22956548 |
| Biological Process | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | ACKR3 | IMP | | Human | PMID:20388803 |
| Biological Process | GO:0001764 | neuron migration | ackr3b | IMP | ZFIN:ZDB-GENO-100507-5 | Zfish | PMID:23382464 |
| Biological Process | GO:0021557 | oculomotor nerve development | ACKR3 | IMP | | Human | PMID:31211835 |
| Biological Process | GO:0021557 | oculomotor nerve development | Ackr3 | IMP | | Mouse | MGI:MGI:6391997|PMID:31211835 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | ACKR3 | IMP | | Human | PMID:22300987 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | Ackr3 | IGI | MGI:MGI:1341264,MGI:MGI:98397 | Mouse | PMID:22956548 |
| Biological Process | GO:0070374 | positive regulation of ERK1 and ERK2 cascade | Ackr3 | IGI | MGI:MGI:1341264,MGI:MGI:95294 | Mouse | PMID:22956548 |
| Biological Process | GO:1905322 | positive regulation of mesenchymal stem cell migration | Ackr3 | IMP | | Rat | PMID:24924806|RGD:11352662 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IMP | ZFIN:ZDB-GENO-100507-5 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IGI | ZFIN:ZDB-MRPHLNO-050318-7,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17570670 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IGI | ZFIN:ZDB-GENO-100507-5,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-100427-5 | Zfish | PMID:17570670 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IGI | ZFIN:ZDB-GENO-030312-1,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17570670 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3b | IGI | ZFIN:ZDB-MRPHLNO-081217-1,ZFIN:ZDB-MRPHLNO-100512-6 | Zfish | PMID:17394634 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3a | IGI | ZFIN:ZDB-GENO-100507-5,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3a | IMP | ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0048920 | posterior lateral line neuromast primordium migration | ackr3a | IGI | ZFIN:ZDB-GENO-140207-11,ZFIN:ZDB-MRPHLNO-140205-5,ZFIN:ZDB-MRPHLNO-140205-6 | Zfish | PMID:24119842 |
| Biological Process | GO:0031623 | receptor internalization | ACKR3 | IMP | | Human | PMID:22300987 |
| Biological Process | GO:0031623 | receptor internalization | ACKR3 | IMP | | Human | PMID:22457824 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | ackr3b | IMP | ZFIN:ZDB-GENO-100507-5 | Zfish | PMID:23382464 |
| Biological Process | GO:0021730 | trigeminal sensory nucleus development | ackr3b | IGI | ZFIN:ZDB-GENO-141204-1 | Zfish | PMID:23382464 |
| Biological Process | GO:0001570 | vasculogenesis | ackr3b | IMP | ZFIN:ZDB-MRPHLNO-071105-1,ZFIN:ZDB-MRPHLNO-071105-2 | Zfish | PMID:17898181 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools