Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Sox7
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes SOX7
- Mouse genes Sox7
- Rat genes Sox7
- Zfish genes sox7
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
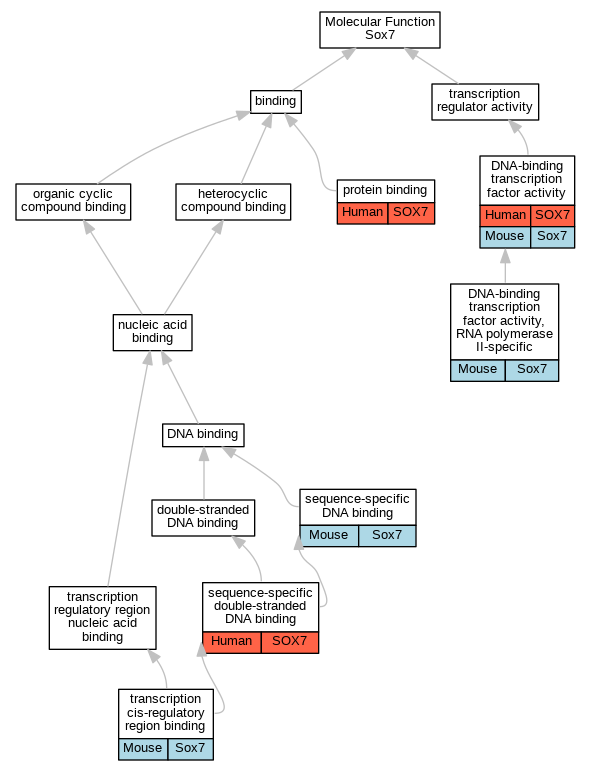
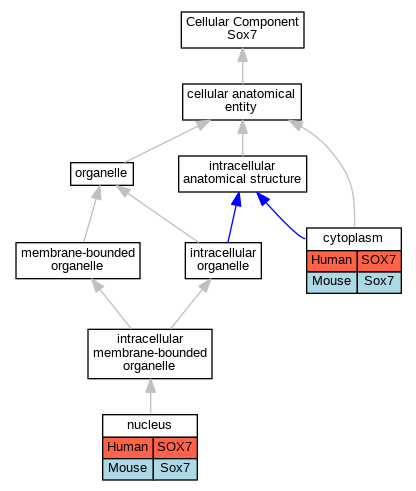
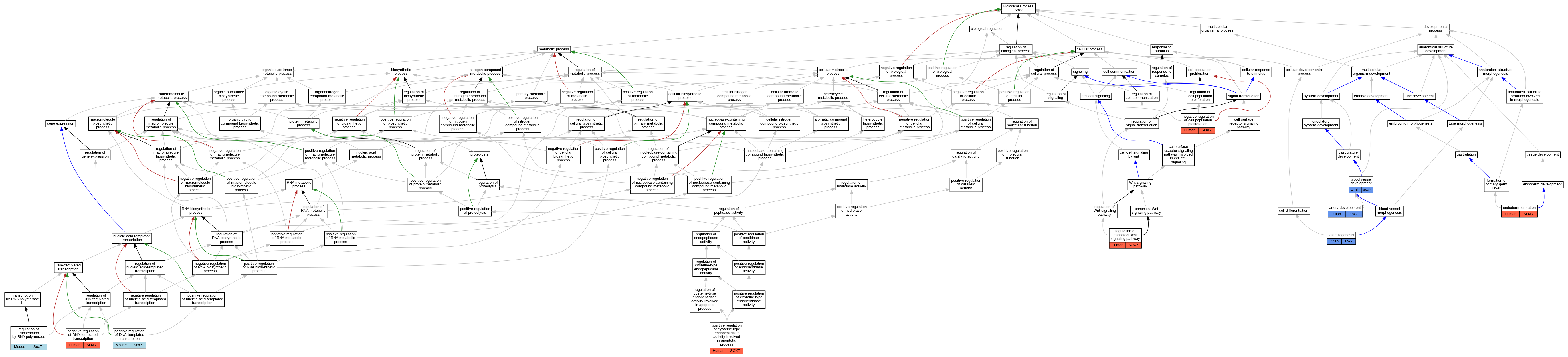 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Sox7
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Sox7
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | SOX7 | IDA | | Human | PMID:19108950 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Sox7 | IDA | | Mouse | PMID:15082719 |
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | Sox7 | IDA | | Mouse | PMID:19515696 |
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | Sox7 | IDA | | Mouse | PMID:15220343 |
| Molecular Function | GO:0005515 | protein binding | SOX7 | IPI | UniProtKB:O76003 | Human | PMID:25416956 |
| Molecular Function | GO:0005515 | protein binding | SOX7 | IPI | UniProtKB:Q8N9R8 | Human | PMID:32296183 |
| Molecular Function | GO:0005515 | protein binding | SOX7 | IPI | UniProtKB:P42858 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | SOX7 | IPI | UniProtKB:Q92870-2 | Human | PMID:32814053 |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Sox7 | IDA | | Mouse | PMID:10320775 |
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | SOX7 | IDA | | Human | PMID:28473536 |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | Sox7 | IDA | | Mouse | MGI:MGI:4887547|PMID:19328208 |
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | Sox7 | IDA | | Mouse | PMID:19515696 |
| Cellular Component | GO:0005737 | cytoplasm | SOX7 | IDA | | Human | PMID:18819930 |
| Cellular Component | GO:0005737 | cytoplasm | Sox7 | IDA | | Mouse | PMID:10320775 |
| Cellular Component | GO:0005634 | nucleus | SOX7 | IDA | | Human | PMID:18682240 |
| Cellular Component | GO:0005634 | nucleus | SOX7 | IDA | | Human | PMID:19108950 |
| Cellular Component | GO:0005634 | nucleus | Sox7 | IDA | | Mouse | PMID:21146513 |
| Cellular Component | GO:0005634 | nucleus | Sox7 | IDA | | Mouse | PMID:10320775 |
| Biological Process | GO:0060840 | artery development | sox7 | IGI | ZFIN:ZDB-MRPHLNO-060626-6,ZFIN:ZDB-MRPHLNO-081022-1,ZFIN:ZDB-MRPHLNO-081022-2 | Zfish | PMID:23818617 |
| Biological Process | GO:0001568 | blood vessel development | sox7 | IMP | ZFIN:ZDB-GENO-150818-2 | Zfish | PMID:25834021 |
| Biological Process | GO:0001568 | blood vessel development | sox7 | IGI | ZFIN:ZDB-GENE-000526-1 | Zfish | PMID:25834021 |
| Biological Process | GO:0001568 | blood vessel development | sox7 | IGI | ZFIN:ZDB-GENE-990415-67 | Zfish | PMID:25834021 |
| Biological Process | GO:0001568 | blood vessel development | sox7 | IGI | ZFIN:ZDB-GENE-080725-1 | Zfish | PMID:25834021 |
| Biological Process | GO:0001706 | endoderm formation | SOX7 | IDA | | Human | PMID:18682240 |
| Biological Process | GO:0008285 | negative regulation of cell population proliferation | SOX7 | IDA | | Human | PMID:19108950 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | SOX7 | IDA | | Human | PMID:19108950 |
| Biological Process | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | SOX7 | IDA | | Human | PMID:19108950 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Sox7 | IDA | | Mouse | PMID:15220343 |
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | Sox7 | IDA | | Mouse | PMID:19515696 |
| Biological Process | GO:0060828 | regulation of canonical Wnt signaling pathway | SOX7 | IDA | | Human | PMID:19108950 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Sox7 | IDA | | Mouse | PMID:15082719 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-081022-3,ZFIN:ZDB-MRPHLNO-081022-4 | Zfish | PMID:18094332 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-080811-1,ZFIN:ZDB-MRPHLNO-080811-3 | Zfish | PMID:18377889 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-080811-2,ZFIN:ZDB-MRPHLNO-080811-4 | Zfish | PMID:18377889 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-081022-1,ZFIN:ZDB-MRPHLNO-081022-2 | Zfish | PMID:21726513 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-081022-1,ZFIN:ZDB-MRPHLNO-081022-2 | Zfish | PMID:18094332 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-080725-1,ZFIN:ZDB-MRPHLNO-080725-2 | Zfish | PMID:18032732 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-081022-1,ZFIN:ZDB-MRPHLNO-081022-2 | Zfish | PMID:21730073 |
| Biological Process | GO:0001570 | vasculogenesis | sox7 | IGI | ZFIN:ZDB-MRPHLNO-081022-5,ZFIN:ZDB-MRPHLNO-081022-6 | Zfish | PMID:18094332 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





