|
Symbol Name ID |
Prdm1
PR domain containing 1, with ZNF domain MGI:99655 |
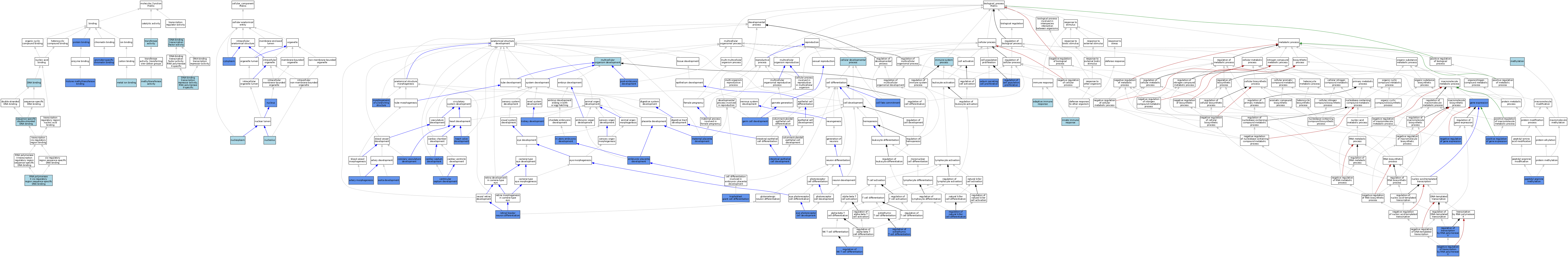
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990226 | histone methyltransferase binding | IPI | J:109626 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008168 | methyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:1990841 | promoter-specific chromatin binding | IDA | J:232474 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:175905 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:109626 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:109626 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:115995 | |||||||||
| Biological Process | GO:0002250 | adaptive immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0035904 | aorta development | IMP | J:222159 | |||||||||
| Biological Process | GO:0048844 | artery morphogenesis | IGI | J:213584 | |||||||||
| Biological Process | GO:0048844 | artery morphogenesis | IMP | J:213584 | |||||||||
| Biological Process | GO:0048844 | artery morphogenesis | IMP | J:213584 | |||||||||
| Biological Process | GO:0003279 | cardiac septum development | IMP | J:222159 | |||||||||
| Biological Process | GO:0045165 | cell fate commitment | IBA | J:265628 | |||||||||
| Biological Process | GO:0045165 | cell fate commitment | IMP | J:101718 | |||||||||
| Biological Process | GO:0045165 | cell fate commitment | IMP | J:99994 | |||||||||
| Biological Process | GO:0048869 | cellular developmental process | IEA | J:72247 | |||||||||
| Biological Process | GO:0060976 | coronary vasculature development | IMP | J:222159 | |||||||||
| Biological Process | GO:0001892 | embryonic placenta development | IMP | J:101718 | |||||||||
| Biological Process | GO:0042462 | eye photoreceptor cell development | IMP | J:160529 | |||||||||
| Biological Process | GO:0042462 | eye photoreceptor cell development | IMP | J:160529 | |||||||||
| Biological Process | GO:0010467 | gene expression | IDA | J:109626 | |||||||||
| Biological Process | GO:0007281 | germ cell development | IGI | J:151610 | |||||||||
| Biological Process | GO:0003170 | heart valve development | IMP | J:222159 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:101718 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0060576 | intestinal epithelial cell development | IMP | J:173543 | |||||||||
| Biological Process | GO:0060576 | intestinal epithelial cell development | IMP | J:173543 | |||||||||
| Biological Process | GO:0001822 | kidney development | IMP | J:231949 | |||||||||
| Biological Process | GO:0001893 | maternal placenta development | IMP | J:101718 | |||||||||
| Biological Process | GO:0032259 | methylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0001763 | morphogenesis of a branching structure | IMP | J:213584 | |||||||||
| Biological Process | GO:0001763 | morphogenesis of a branching structure | IMP | J:213584 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | IMP | J:173543 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | IMP | J:173543 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:160529 | |||||||||
| Biological Process | GO:0018216 | peptidyl-arginine methylation | IDA | J:109626 | |||||||||
| Biological Process | GO:0010628 | positive regulation of gene expression | IDA | J:138571 | |||||||||
| Biological Process | GO:0009791 | post-embryonic development | IMP | J:173543 | |||||||||
| Biological Process | GO:0009791 | post-embryonic development | IMP | J:173543 | |||||||||
| Biological Process | GO:0042127 | regulation of cell population proliferation | IMP | J:213584 | |||||||||
| Biological Process | GO:0042127 | regulation of cell population proliferation | IMP | J:213584 | |||||||||
| Biological Process | GO:0033082 | regulation of extrathymic T cell differentiation | IMP | J:232474 | |||||||||
| Biological Process | GO:0032823 | regulation of natural killer cell differentiation | IMP | J:232474 | |||||||||
| Biological Process | GO:0051136 | regulation of NK T cell differentiation | IMP | J:232474 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IMP | J:240687 | |||||||||
| Biological Process | GO:0060040 | retinal bipolar neuron differentiation | IMP | J:240687 | |||||||||
| Biological Process | GO:1990654 | sebum secreting cell proliferation | IDA | J:115995 | |||||||||
| Biological Process | GO:0060707 | trophoblast giant cell differentiation | IMP | J:187742 | |||||||||
| Biological Process | GO:0003281 | ventricular septum development | IMP | J:222159 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||