|
Symbol Name ID |
Clspn
claspin MGI:2445153 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
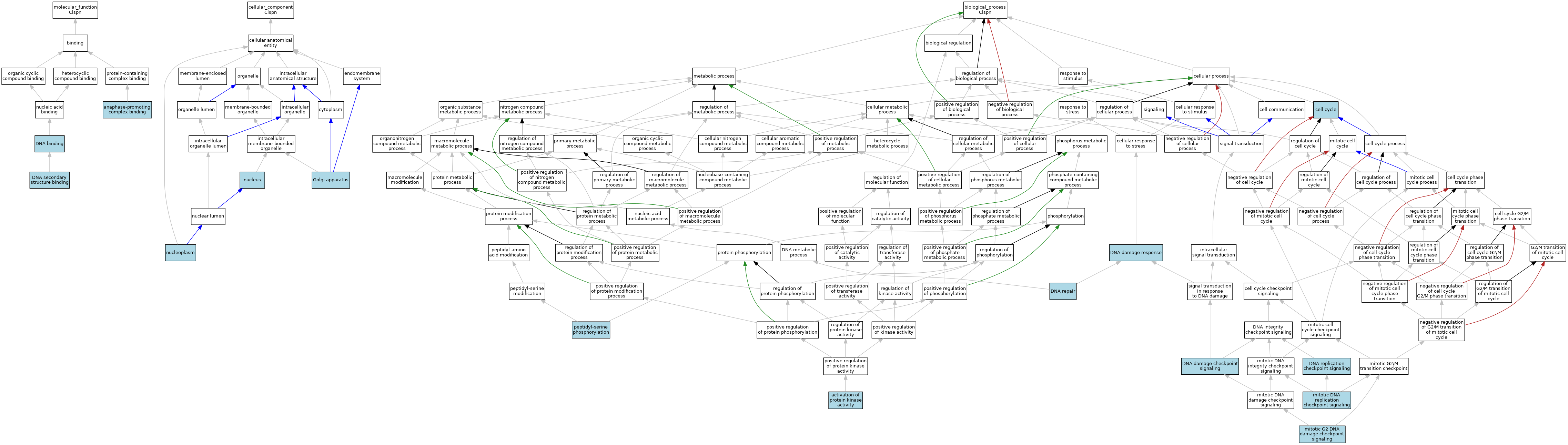
| Molecular Function | GO:0010997 | anaphase-promoting complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0010997 | anaphase-promoting complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000217 | DNA secondary structure binding | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:73065 | |||||||||
| Biological Process | GO:0032147 | activation of protein kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0032147 | activation of protein kinase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0000077 | DNA damage checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0000076 | DNA replication checkpoint signaling | ISO | J:73065 | |||||||||
| Biological Process | GO:0033314 | mitotic DNA replication checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0033314 | mitotic DNA replication checkpoint signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0007095 | mitotic G2 DNA damage checkpoint signaling | IBA | J:265628 | |||||||||
| Biological Process | GO:0018105 | peptidyl-serine phosphorylation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||