|
Symbol Name ID |
Irf7
interferon regulatory factor 7 MGI:1859212 |
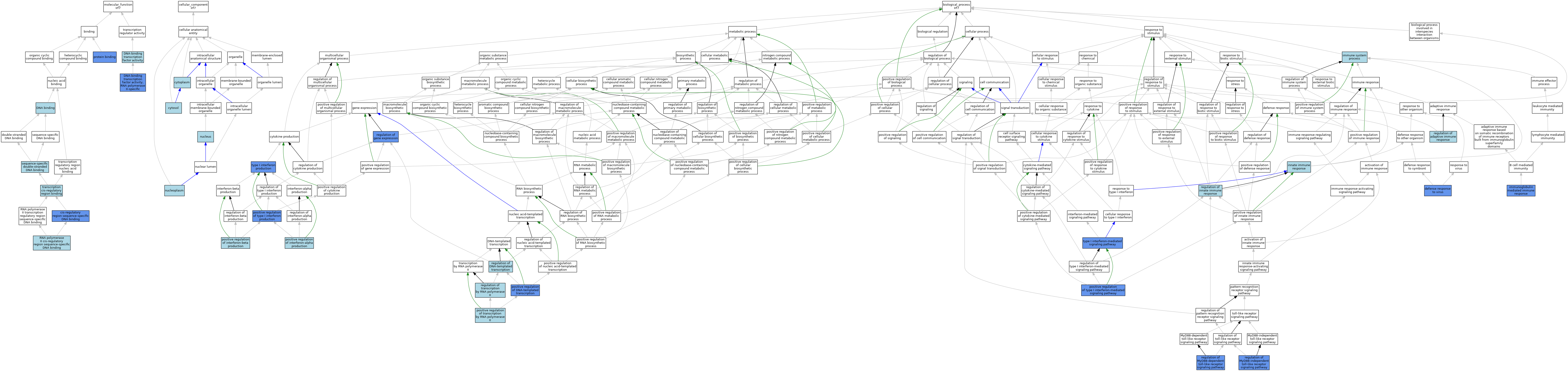
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0000987 | cis-regulatory region sequence-specific DNA binding | IDA | J:235021 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IMP | J:235021 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IDA | J:188556 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:107628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:221417 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:102933 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:180378 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:205872 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000976 | transcription cis-regulatory region binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0051607 | defense response to virus | ISO | J:164563 | |||||||||
| Biological Process | GO:0051607 | defense response to virus | IMP | J:235021 | |||||||||
| Biological Process | GO:0002376 | immune system process | IBA | J:265628 | |||||||||
| Biological Process | GO:0016064 | immunoglobulin mediated immune response | IMP | J:97662 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | IMP | J:235021 | |||||||||
| Biological Process | GO:0032727 | positive regulation of interferon-alpha production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032727 | positive regulation of interferon-alpha production | TAS | J:184127 | |||||||||
| Biological Process | GO:0032728 | positive regulation of interferon-beta production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032728 | positive regulation of interferon-beta production | TAS | J:184127 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0032481 | positive regulation of type I interferon production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032481 | positive regulation of type I interferon production | IGI | J:205872 | |||||||||
| Biological Process | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway | IMP | J:123411 | |||||||||
| Biological Process | GO:0002819 | regulation of adaptive immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IGI | J:191073 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IGI | J:191073 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IGI | J:191073 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | IGI | J:191073 | |||||||||
| Biological Process | GO:0045088 | regulation of innate immune response | TAS | J:97662 | |||||||||
| Biological Process | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway | IMP | J:97662 | |||||||||
| Biological Process | GO:0034127 | regulation of MyD88-independent toll-like receptor signaling pathway | IMP | J:97662 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0032606 | type I interferon production | IMP | J:97662 | |||||||||
| Biological Process | GO:0060337 | type I interferon-mediated signaling pathway | IMP | J:235021 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||