|
Symbol Name ID |
Ddb1
damage specific DNA binding protein 1 MGI:1202384 |
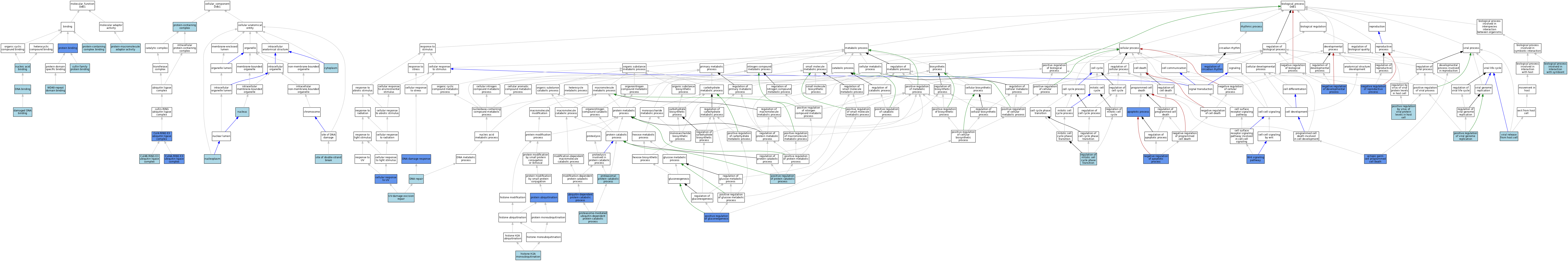
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0097602 | cullin family protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:331350 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:266543 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:283196 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:245381 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222132 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030674 | protein-macromolecule adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0071987 | WD40-repeat domain binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex | IDA | J:302674 | |||||||||
| Cellular Component | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex | EXP | J:320034 | |||||||||
| Cellular Component | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex | ISO | J:73065 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | NAS | J:320034 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | IBA | J:265628 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:205485 | |||||||||
| Biological Process | GO:0051702 | biological process involved in interaction with symbiont | ISO | J:164563 | |||||||||
| Biological Process | GO:0034644 | cellular response to UV | ISO | J:164563 | |||||||||
| Biological Process | GO:0034644 | cellular response to UV | EXP | J:320034 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | EXP | J:320034 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IBA | J:265628 | |||||||||
| Biological Process | GO:0035234 | ectopic germ cell programmed cell death | IMP | J:205485 | |||||||||
| Biological Process | GO:0035518 | histone H2A monoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:205485 | |||||||||
| Biological Process | GO:0051093 | negative regulation of developmental process | IMP | J:205485 | |||||||||
| Biological Process | GO:2000242 | negative regulation of reproductive process | IMP | J:205485 | |||||||||
| Biological Process | GO:0046726 | positive regulation by virus of viral protein levels in host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0045722 | positive regulation of gluconeogenesis | IMP | J:245577 | |||||||||
| Biological Process | GO:0045732 | positive regulation of protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0045070 | positive regulation of viral genome replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0010498 | proteasomal protein catabolic process | ISO | J:235324 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | IMP | J:243464 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0042752 | regulation of circadian rhythm | IMP | J:243464 | |||||||||
| Biological Process | GO:1901990 | regulation of mitotic cell cycle phase transition | ISO | J:164563 | |||||||||
| Biological Process | GO:0048511 | rhythmic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IMP | J:243464 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0070914 | UV-damage excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0019076 | viral release from host cell | ISO | J:164563 | |||||||||
| Biological Process | GO:0016055 | Wnt signaling pathway | IDA | J:142037 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||