|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
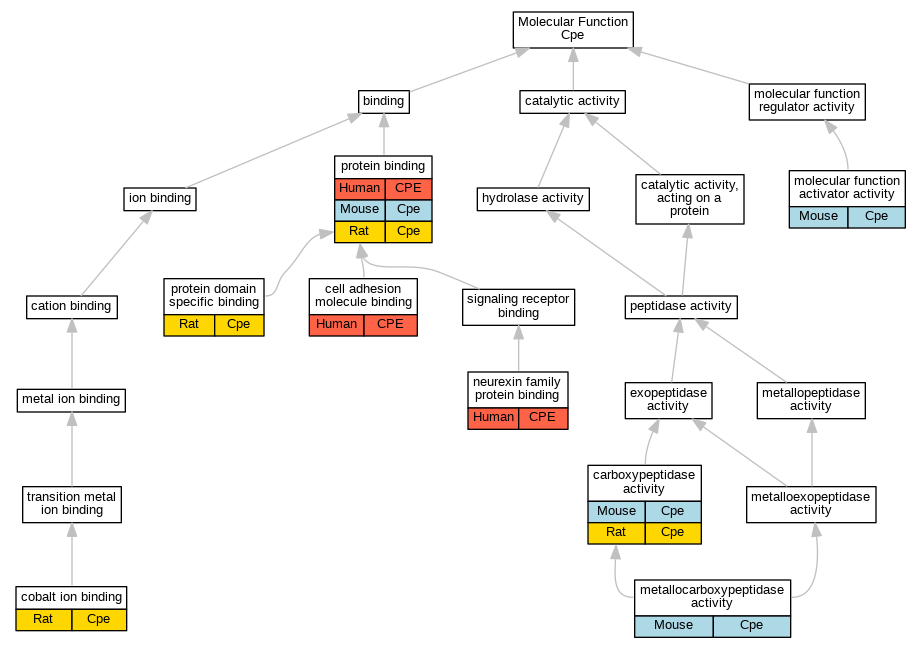
| Molecular Function | GO:0004180 | carboxypeptidase activity | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:7663508 |
| Molecular Function | GO:0004180 | carboxypeptidase activity | Cpe | IDA | Rat | PMID:2822027|RGD:1626225 | |
| Molecular Function | GO:0050839 | cell adhesion molecule binding | CPE | IPI | UniProtKB:Q9UHC6 | Human | PMID:19166515 |
| Molecular Function | GO:0050897 | cobalt ion binding | Cpe | IMP | Rat | PMID:2822027|RGD:1626225 | |
| Molecular Function | GO:0004181 | metallocarboxypeptidase activity | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:9348219 |
| Molecular Function | GO:0140677 | molecular function activator activity | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:9348219 |
| Molecular Function | GO:0042043 | neurexin family protein binding | CPE | IPI | UniProtKB:Q9UHC6 | Human | PMID:19166515 |
| Molecular Function | GO:0005515 | protein binding | CPE | IPI | UniProtKB:P49768-2 | Human | PMID:32814053 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | PR:P01193 | Mouse | PMID:16219686 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | PR:P47867 | Mouse | PMID:16219686 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | RGD:1349318 | Rat | PMID:15233624|RGD:6483328 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | RGD:732004 | Rat | PMID:16219686|RGD:1626212 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | RGD:1307076 | Rat | PMID:19166515|RGD:6483326 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | RGD:3715 | Rat | PMID:19419578|RGD:6483104 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | RGD:620106 | Rat | PMID:19553464|RGD:6483099 |
| Molecular Function | GO:0005515 | protein binding | Cpe | IPI | UniProtKB:P02866 | Rat | PMID:2822027|RGD:1626225 |
| Molecular Function | GO:0019904 | protein domain specific binding | Cpe | IDA | Rat | PMID:19553464|RGD:6483099 | |
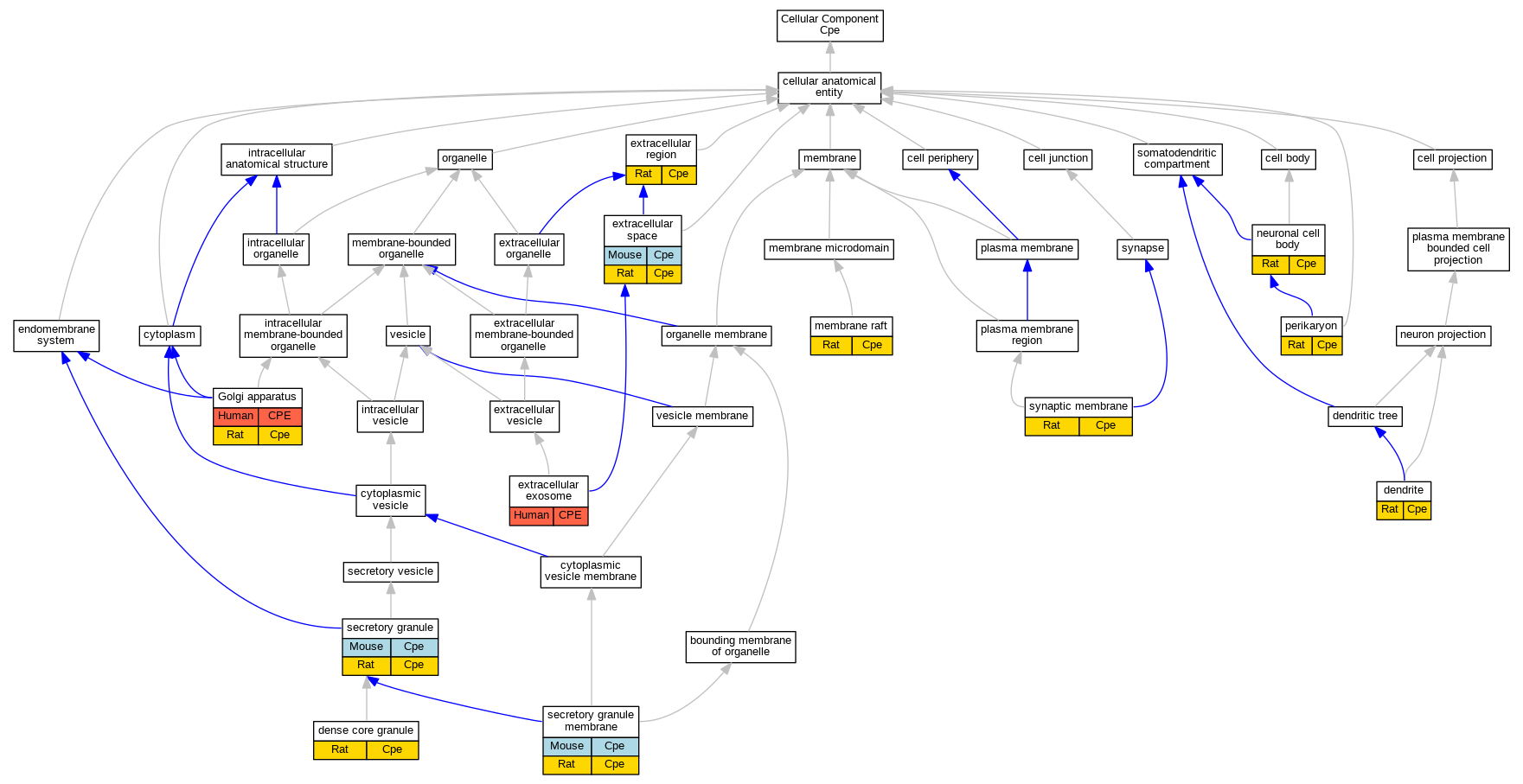
| Cellular Component | GO:0030425 | dendrite | Cpe | IDA | Rat | PMID:17543468|RGD:2308893 | |
| Cellular Component | GO:0030425 | dendrite | Cpe | IDA | Rat | PMID:19166515|RGD:6483326 | |
| Cellular Component | GO:0031045 | dense core granule | Cpe | IDA | Rat | PMID:19553464|RGD:6483099 | |
| Cellular Component | GO:0070062 | extracellular exosome | CPE | HDA | Human | PMID:23533145 | |
| Cellular Component | GO:0005576 | extracellular region | Cpe | IDA | Rat | PMID:10386951|RGD:6483348 | |
| Cellular Component | GO:0005615 | extracellular space | Cpe | HDA | Mouse | MGI:MGI:5640941|PMID:24006456 | |
| Cellular Component | GO:0005615 | extracellular space | Cpe | IDA | Rat | PMID:10206965|RGD:1626217 | |
| Cellular Component | GO:0005794 | Golgi apparatus | CPE | IDA | Human | PMID:19166515 | |
| Cellular Component | GO:0005794 | Golgi apparatus | Cpe | IDA | Rat | PMID:19166515|RGD:6483326 | |
| Cellular Component | GO:0005794 | Golgi apparatus | Cpe | IDA | Rat | PMID:9173892|RGD:1626218 | |
| Cellular Component | GO:0045121 | membrane raft | Cpe | IDA | Rat | PMID:14597614|RGD:6483330 | |
| Cellular Component | GO:0043025 | neuronal cell body | Cpe | IDA | Rat | PMID:17543468|RGD:2308893 | |
| Cellular Component | GO:0043204 | perikaryon | Cpe | IDA | Rat | PMID:17543468|RGD:2308893 | |
| Cellular Component | GO:0043204 | perikaryon | Cpe | IDA | Rat | PMID:19166515|RGD:6483326 | |
| Cellular Component | GO:0030141 | secretory granule | Cpe | IDA | Mouse | PMID:9348219 | |
| Cellular Component | GO:0030141 | secretory granule | Cpe | IDA | Rat | PMID:2822027|RGD:1626225 | |
| Cellular Component | GO:0030667 | secretory granule membrane | Cpe | IDA | Mouse | PMID:16219686 | |
| Cellular Component | GO:0030667 | secretory granule membrane | Cpe | IDA | Rat | PMID:14597614|RGD:6483330 | |
| Cellular Component | GO:0097060 | synaptic membrane | Cpe | IDA | Rat | PMID:19553464|RGD:6483099 | |
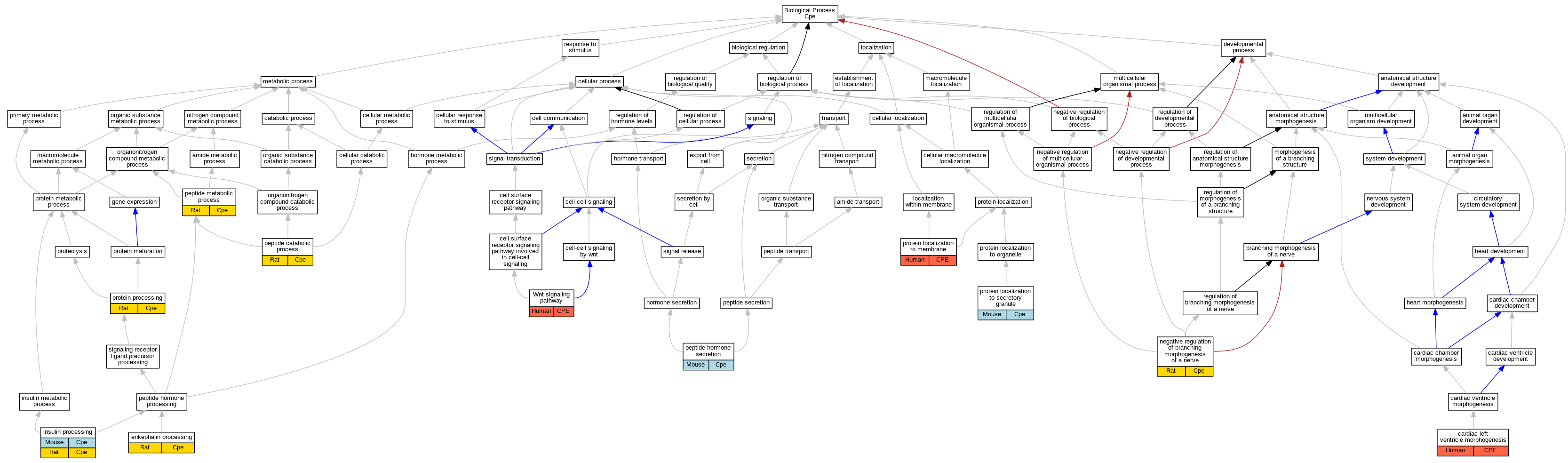
| Biological Process | GO:0003214 | cardiac left ventricle morphogenesis | CPE | IMP | Human | PMID:19593212 | |
| Biological Process | GO:0034230 | enkephalin processing | Cpe | IDA | Rat | PMID:6326144|RGD:6483103 | |
| Biological Process | GO:0030070 | insulin processing | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:9348219 |
| Biological Process | GO:0030070 | insulin processing | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:7663508 |
| Biological Process | GO:0030070 | insulin processing | Cpe | IDA | Rat | PMID:2822027|RGD:1626225 | |
| Biological Process | GO:2000173 | negative regulation of branching morphogenesis of a nerve | Cpe | IMP | Rat | PMID:19553464|RGD:6483099 | |
| Biological Process | GO:0043171 | peptide catabolic process | Cpe | IDA | Rat | PMID:2822027|RGD:1626225 | |
| Biological Process | GO:0030072 | peptide hormone secretion | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:16219686 |
| Biological Process | GO:0006518 | peptide metabolic process | Cpe | IDA | Rat | PMID:16219686|RGD:1626212 | |
| Biological Process | GO:0072657 | protein localization to membrane | CPE | IDA | Human | PMID:19166515 | |
| Biological Process | GO:0033366 | protein localization to secretory granule | Cpe | IMP | MGI:MGI:1856395 | Mouse | PMID:16219686 |
| Biological Process | GO:0016485 | protein processing | Cpe | IDA | Rat | PMID:2822027|RGD:1626225 | |
| Biological Process | GO:0016055 | Wnt signaling pathway | CPE | IDA | Human | PMID:22824791 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||