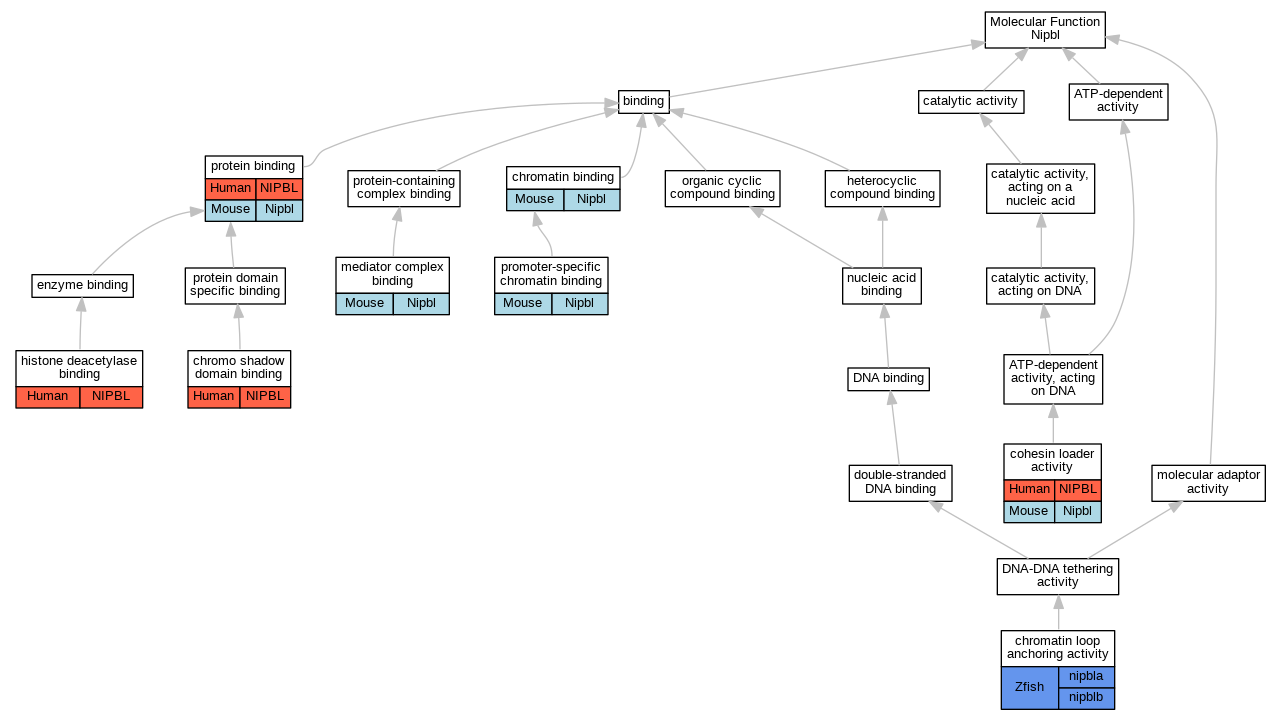
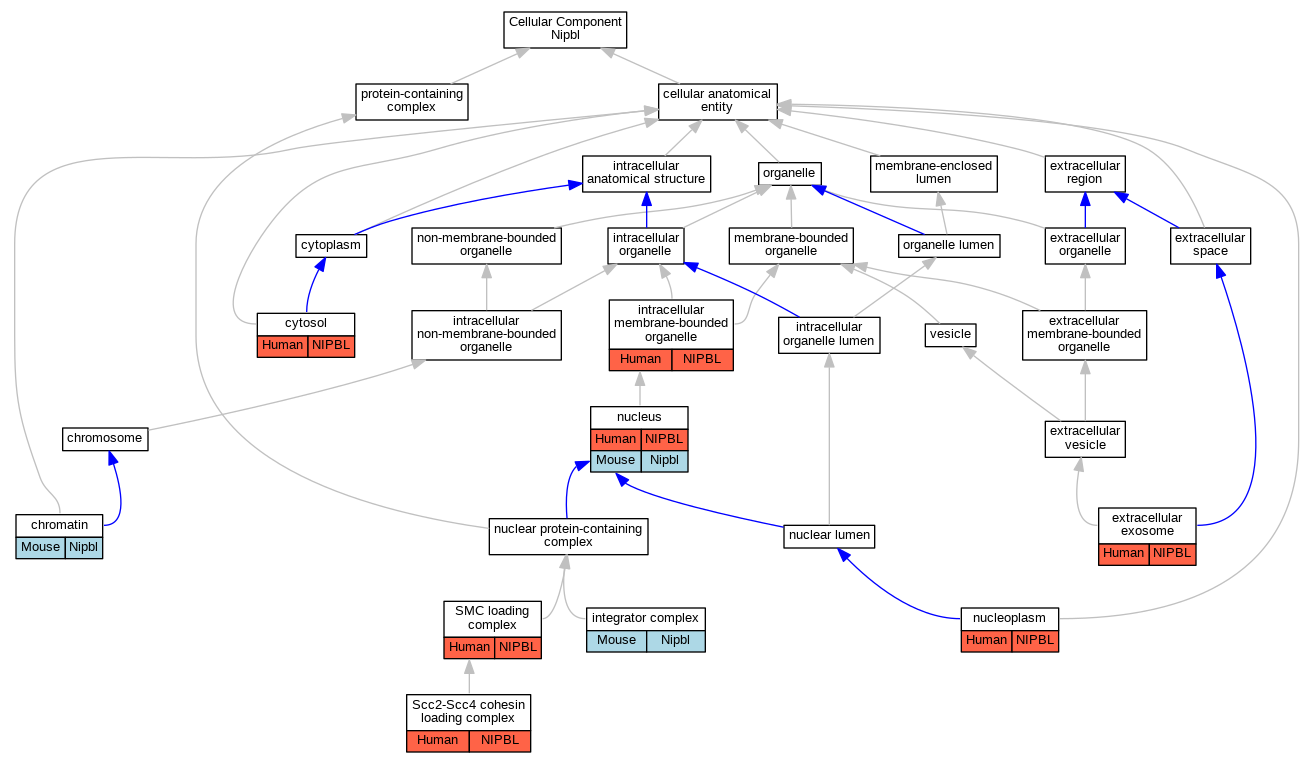
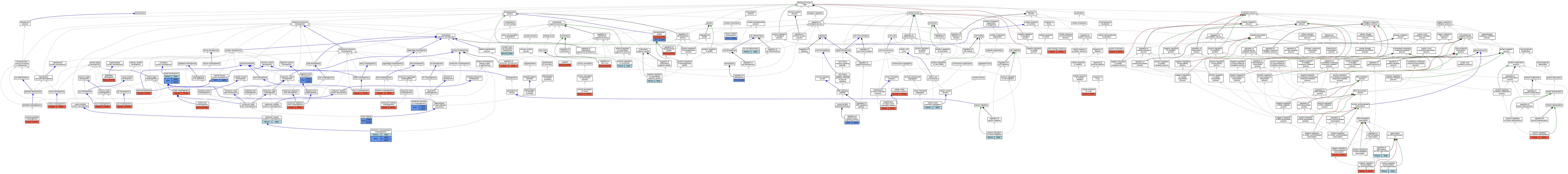
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0003682 | chromatin binding | Nipbl | IDA | | Mouse | PMID:20720539 |
| Molecular Function | GO:0140587 | chromatin loop anchoring activity | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Molecular Function | GO:0140587 | chromatin loop anchoring activity | nipblb | IGI | ZFIN:ZDB-MRPHLNO-080819-1,ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Molecular Function | GO:0140587 | chromatin loop anchoring activity | nipbla | IGI | ZFIN:ZDB-MRPHLNO-080819-1,ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Molecular Function | GO:0140587 | chromatin loop anchoring activity | nipbla | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Molecular Function | GO:0070087 | chromo shadow domain binding | NIPBL | IPI | UniProtKB:P45973 | Human | PMID:15882967 |
| Molecular Function | GO:0061775 | cohesin loader activity | NIPBL | IMP | | Human | PMID:22628566 |
| Molecular Function | GO:0061775 | cohesin loader activity | Nipbl | IMP | | Mouse | MGI:MGI:6102971|PMID:29094699 |
| Molecular Function | GO:0042826 | histone deacetylase binding | NIPBL | IPI | UniProtKB:O15379 | Human | PMID:18854353 |
| Molecular Function | GO:0042826 | histone deacetylase binding | NIPBL | IPI | UniProtKB:Q13547 | Human | PMID:18854353 |
| Molecular Function | GO:0036033 | mediator complex binding | Nipbl | IDA | | Mouse | PMID:20720539 |
| Molecular Function | GO:1990841 | promoter-specific chromatin binding | Nipbl | IDA | | Mouse | MGI:MGI:5908762|PMID:28041881 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9Y6X3 | Human | PMID:16682347 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9Y6X3 | Human | PMID:16802858 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9BQA5 | Human | PMID:17577209 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9Y6X3 | Human | PMID:21934712 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9Y6X3 | Human | PMID:22628566 |
| Molecular Function | GO:0005515 | protein binding | NIPBL | IPI | UniProtKB:Q9Y6X3 | Human | PMID:26496610 |
| Molecular Function | GO:0005515 | protein binding | Nipbl | IPI | UniProtKB:Q8BZ47 | Mouse | MGI:MGI:5908762|PMID:28041881 |
| Cellular Component | GO:0000785 | chromatin | Nipbl | IDA | | Mouse | PMID:23920377 |
| Cellular Component | GO:0005829 | cytosol | NIPBL | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0070062 | extracellular exosome | NIPBL | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0032039 | integrator complex | Nipbl | IDA | | Mouse | MGI:MGI:5908762|PMID:28041881 |
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | NIPBL | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005654 | nucleoplasm | NIPBL | IDA | | Human | GO_REF:0000052 |
| Cellular Component | GO:0005634 | nucleus | NIPBL | IDA | | Human | PMID:16802858 |
| Cellular Component | GO:0005634 | nucleus | Nipbl | IDA | | Mouse | MGI:MGI:6102971|PMID:29094699 |
| Cellular Component | GO:0090694 | Scc2-Scc4 cohesin loading complex | NIPBL | IDA | | Human | PMID:22628566 |
| Cellular Component | GO:0032116 | SMC loading complex | NIPBL | IDA | | Human | PMID:16682347 |
| Biological Process | GO:0007420 | brain development | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0071481 | cellular response to X-ray | NIPBL | IMP | | Human | PMID:17468178 |
| Biological Process | GO:0007417 | central nervous system development | nipblb | IMP | ZFIN:ZDB-MRPHLNO-131219-3 | Zfish | PMID:24136230 |
| Biological Process | GO:0050890 | cognition | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0048589 | developmental growth | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0048589 | developmental growth | nipblb | IGI | ZFIN:ZDB-MRPHLNO-131219-3,ZFIN:ZDB-MRPHLNO-160407-3 | Zfish | PMID:26206533 |
| Biological Process | GO:0048565 | digestive tract development | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0048565 | digestive tract development | nipbla | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0006974 | DNA damage response | NIPBL | IMP | | Human | PMID:17468178 |
| Biological Process | GO:0042471 | ear morphogenesis | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0048701 | embryonic cranial skeleton morphogenesis | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:0048557 | embryonic digestive tract morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0048557 | embryonic digestive tract morphogenesis | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0035115 | embryonic forelimb morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0035115 | embryonic forelimb morphogenesis | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0035118 | embryonic pectoral fin morphogenesis | nipblb | IGI | ZFIN:ZDB-MRPHLNO-080819-1,ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Biological Process | GO:0035118 | embryonic pectoral fin morphogenesis | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Biological Process | GO:0035118 | embryonic pectoral fin morphogenesis | nipbla | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Biological Process | GO:0035118 | embryonic pectoral fin morphogenesis | nipbla | IGI | ZFIN:ZDB-MRPHLNO-080819-1,ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25255084 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25378554 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | nipbla | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:25378554 |
| Biological Process | GO:0035261 | external genitalia morphogenesis | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0035261 | external genitalia morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0048592 | eye morphogenesis | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0060325 | face morphogenesis | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0060325 | face morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0045444 | fat cell differentiation | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:0035136 | forelimb morphogenesis | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0061010 | gallbladder development | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0007507 | heart development | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:0007507 | heart development | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0007507 | heart development | nipbla | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0007507 | heart development | nipbla | IMP | ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0003146 | heart jogging | nipblb | IGI | ZFIN:ZDB-MRPHLNO-111201-1,ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0003146 | heart jogging | nipbla | IMP | ZFIN:ZDB-MRPHLNO-111201-3 | Zfish | PMID:22039349 |
| Biological Process | GO:0003007 | heart morphogenesis | NIPBL | IMP | | Human | PMID:8291537 |
| Biological Process | GO:0034088 | maintenance of mitotic sister chromatid cohesion | NIPBL | IMP | | Human | PMID:16682347 |
| Biological Process | GO:0007064 | mitotic sister chromatid cohesion | NIPBL | IMP | | Human | PMID:16100726 |
| Biological Process | GO:0007064 | mitotic sister chromatid cohesion | NIPBL | IDA | | Human | PMID:22628566 |
| Biological Process | GO:0000070 | mitotic sister chromatid segregation | Nipbl | IMP | | Mouse | MGI:MGI:6102971|PMID:29094699 |
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | NIPBL | IDA | | Human | PMID:18854353 |
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | NIPBL | IDA | | Human | PMID:18854353 |
| Biological Process | GO:0070050 | neuron cellular homeostasis | nipblb | IMP | ZFIN:ZDB-MRPHLNO-131219-3 | Zfish | PMID:26206533 |
| Biological Process | GO:0003151 | outflow tract morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0031065 | positive regulation of histone deacetylation | NIPBL | IDA | | Human | PMID:18854353 |
| Biological Process | GO:0040018 | positive regulation of multicellular organism growth | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:2001224 | positive regulation of neuron migration | Nipbl | IMP | | Mouse | MGI:MGI:5908762|PMID:28041881 |
| Biological Process | GO:0045778 | positive regulation of ossification | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:19763162 |
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | Nipbl | IMP | MGI:MGI:4332250 | Mouse | PMID:23920377 |
| Biological Process | GO:0008104 | protein localization | NIPBL | IMP | | Human | PMID:17468178 |
| Biological Process | GO:0060828 | regulation of canonical Wnt signaling pathway | nipblb | IMP | ZFIN:ZDB-MRPHLNO-131219-3 | Zfish | PMID:24136230 |
| Biological Process | GO:0048638 | regulation of developmental growth | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0045995 | regulation of embryonic development | NIPBL | IMP | | Human | PMID:19242925 |
| Biological Process | GO:0042634 | regulation of hair cycle | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:1903706 | regulation of hemopoiesis | nipblb | IMP | ZFIN:ZDB-MRPHLNO-131219-3 | Zfish | PMID:32323916 |
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | Nipbl | IMP | | Mouse | MGI:MGI:5908762|PMID:28041881 |
| Biological Process | GO:0007605 | sensory perception of sound | NIPBL | IMP | | Human | PMID:15146186 |
| Biological Process | GO:0035019 | somatic stem cell population maintenance | Nipbl | IMP | | Mouse | PMID:20720539 |
| Biological Process | GO:0061038 | uterus morphogenesis | NIPBL | IMP | | Human | PMID:19242925 |
 Analysis Tools
Analysis Tools