|
Symbol Name ID |
Plscr1
phospholipid scramblase 1 MGI:893575 |
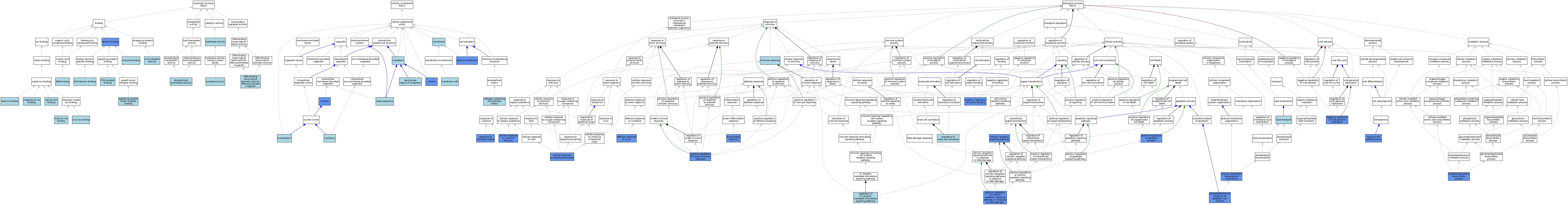
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005509 | calcium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042609 | CD4 receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005154 | epidermal growth factor receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0032791 | lead ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000287 | magnesium ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0045340 | mercury ion binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017128 | phospholipid scramblase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017128 | phospholipid scramblase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:292529 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0001618 | virus receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:127023 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:127023 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:175205 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IBA | J:265628 | |||||||||
| Biological Process | GO:0006953 | acute-phase response | IEP | J:127023 | |||||||||
| Biological Process | GO:0071345 | cellular response to cytokine stimulus | IEP | J:127023 | |||||||||
| Biological Process | GO:0071222 | cellular response to lipopolysaccharide | IEP | J:127023 | |||||||||
| Biological Process | GO:0051607 | defense response to virus | IMP | J:175192 | |||||||||
| Biological Process | GO:0006955 | immune response | ISO | J:155856 | |||||||||
| Biological Process | GO:0097193 | intrinsic apoptotic signaling pathway | IDA | J:182395 | |||||||||
| Biological Process | GO:0006869 | lipid transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0030099 | myeloid cell differentiation | IMP | J:92404 | |||||||||
| Biological Process | GO:0032091 | negative regulation of protein binding | IDA | J:108434 | |||||||||
| Biological Process | GO:0045071 | negative regulation of viral genome replication | IMP | J:175192 | |||||||||
| Biological Process | GO:0006659 | phosphatidylserine biosynthetic process | IDA | J:175205 | |||||||||
| Biological Process | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | IMP | J:292529 | |||||||||
| Biological Process | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | IBA | J:265628 | |||||||||
| Biological Process | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface | IDA | J:175205 | |||||||||
| Biological Process | GO:0017121 | plasma membrane phospholipid scrambling | ISO | J:155856 | |||||||||
| Biological Process | GO:0017121 | plasma membrane phospholipid scrambling | IDA | J:175205 | |||||||||
| Biological Process | GO:0043065 | positive regulation of apoptotic process | IGI | J:108434 | |||||||||
| Biological Process | GO:0045089 | positive regulation of innate immune response | IMP | J:175192 | |||||||||
| Biological Process | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage | IDA | J:175205 | |||||||||
| Biological Process | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0033003 | regulation of mast cell activation | ISO | J:155856 | |||||||||
| Biological Process | GO:0035455 | response to interferon-alpha | IMP | J:175192 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||