|
Symbol Name ID |
Ubqln4
ubiquilin 4 MGI:2150152 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
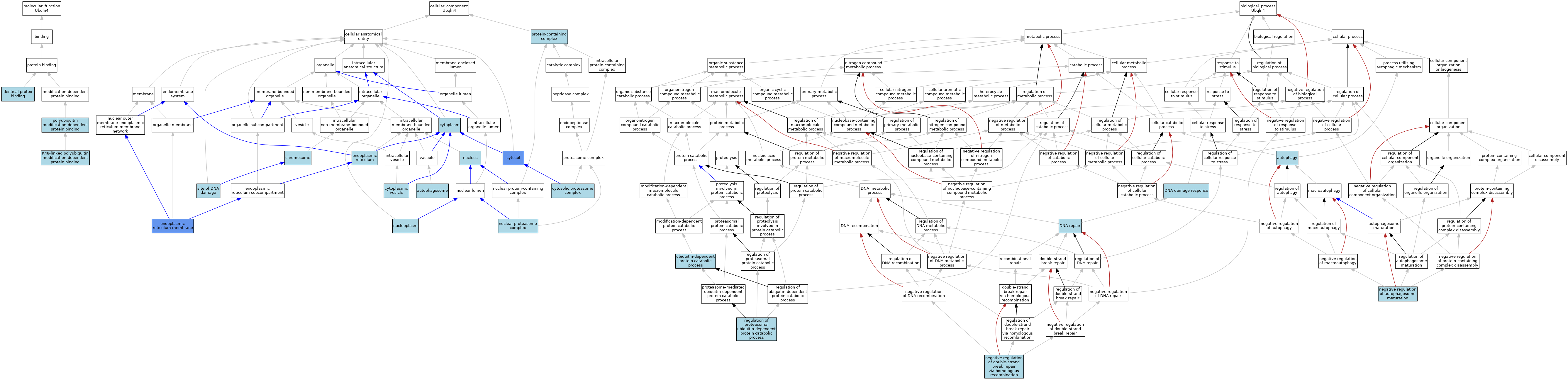
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0036435 | K48-linked polyubiquitin modification-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005776 | autophagosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:70982 | |||||||||
| Cellular Component | GO:0031597 | cytosolic proteasome complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005789 | endoplasmic reticulum membrane | IDA | J:70982 | |||||||||
| Cellular Component | GO:0031595 | nuclear proteasome complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0090734 | site of DNA damage | ISO | J:164563 | |||||||||
| Biological Process | GO:0006914 | autophagy | IEA | J:60000 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:1901097 | negative regulation of autophagosome maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:2000042 | negative regulation of double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||