|
Symbol Name ID |
Xpo1
exportin 1 MGI:2144013 |
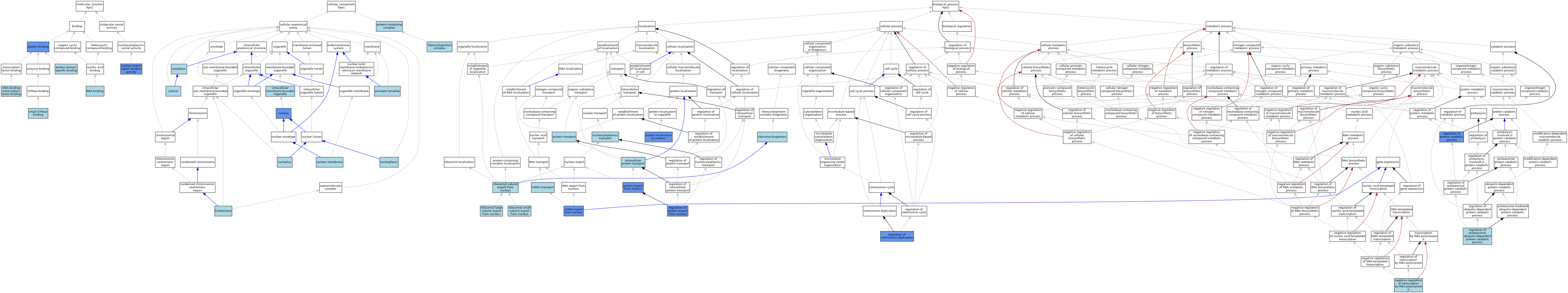
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | IMP | J:285490 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | IMP | J:80612 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | IDA | J:285490 | |||||||||
| Molecular Function | GO:0005049 | nuclear export signal receptor activity | IDA | J:80612 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:247413 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:196265 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:222157 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:248238 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:247411 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:231233 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:97559 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:193640 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005642 | annulate lamellae | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000776 | kinetochore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:103773 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990904 | ribonucleoprotein complex | ISO | J:142821 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | IEA | J:72247 | |||||||||
| Biological Process | GO:0006406 | mRNA export from nucleus | IMP | J:285490 | |||||||||
| Biological Process | GO:0006406 | mRNA export from nucleus | IDA | J:285490 | |||||||||
| Biological Process | GO:0051028 | mRNA transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:155856 | |||||||||
| Biological Process | GO:0006913 | nucleocytoplasmic transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | ISO | J:155856 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IDA | J:142024 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:103171 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IDA | J:75684 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:106641 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:107024 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:97559 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IDA | J:80612 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:137074 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | ISO | J:73065 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:154728 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:148303 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:153534 | |||||||||
| Biological Process | GO:0006611 | protein export from nucleus | IMP | J:105832 | |||||||||
| Biological Process | GO:0034504 | protein localization to nucleus | IMP | J:103773 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0010824 | regulation of centrosome duplication | IMP | J:103773 | |||||||||
| Biological Process | GO:0032434 | regulation of proteasomal ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0042176 | regulation of protein catabolic process | IMP | J:97559 | |||||||||
| Biological Process | GO:0046825 | regulation of protein export from nucleus | IMP | J:88659 | |||||||||
| Biological Process | GO:0000055 | ribosomal large subunit export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0000055 | ribosomal large subunit export from nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0000056 | ribosomal small subunit export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0000056 | ribosomal small subunit export from nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0000054 | ribosomal subunit export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0042254 | ribosome biogenesis | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||