|
Symbol Name ID |
Hdac9
histone deacetylase 9 MGI:1931221 |
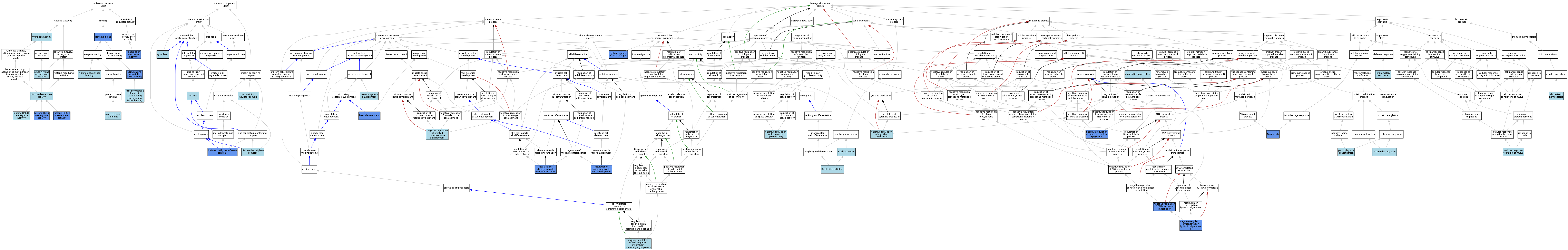
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | IPI | J:152666 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0140297 | DNA-binding transcription factor binding | TAS | J:113927 | |||||||||
| Molecular Function | GO:0004407 | histone deacetylase activity | TAS | J:113927 | |||||||||
| Molecular Function | GO:0042826 | histone deacetylase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0042826 | histone deacetylase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031078 | histone H3K14 deacetylase activity | IMP | J:175911 | |||||||||
| Molecular Function | GO:0032129 | histone H3K9 deacetylase activity | IMP | J:175911 | |||||||||
| Molecular Function | GO:0034739 | histone H4K16 deacetylase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162637 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114108 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:279931 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:70040 | |||||||||
| Molecular Function | GO:0005080 | protein kinase C binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0033558 | protein lysine deacetylase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IDA | J:67665 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | TAS | J:113927 | |||||||||
| Cellular Component | GO:0000118 | histone deacetylase complex | TAS | J:113927 | |||||||||
| Cellular Component | GO:0035097 | histone methyltransferase complex | IDA | J:152666 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | TAS | J:113927 | |||||||||
| Cellular Component | GO:0005667 | transcription regulator complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0042113 | B cell activation | TAS | J:113927 | |||||||||
| Biological Process | GO:0030183 | B cell differentiation | TAS | J:113927 | |||||||||
| Biological Process | GO:0032869 | cellular response to insulin stimulus | ISO | J:164563 | |||||||||
| Biological Process | GO:0042632 | cholesterol homeostasis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | TAS | J:113927 | |||||||||
| Biological Process | GO:0008340 | determination of adult lifespan | IGI | J:174530 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IGI | J:174530 | |||||||||
| Biological Process | GO:0007507 | heart development | IGI | J:93021 | |||||||||
| Biological Process | GO:0016575 | histone deacetylation | IEA | J:72247 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | TAS | J:113927 | |||||||||
| Biological Process | GO:0001818 | negative regulation of cytokine production | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | TAS | J:113927 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IDA | J:67665 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IDA | J:102395 | |||||||||
| Biological Process | GO:0045814 | negative regulation of gene expression, epigenetic | IMP | J:175911 | |||||||||
| Biological Process | GO:0045814 | negative regulation of gene expression, epigenetic | ISO | J:164563 | |||||||||
| Biological Process | GO:0051005 | negative regulation of lipoprotein lipase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0045843 | negative regulation of striated muscle tissue development | TAS | J:113927 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:73065 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IMP | J:107465 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:70040 | |||||||||
| Biological Process | GO:0007399 | nervous system development | TAS | J:113927 | |||||||||
| Biological Process | GO:0034983 | peptidyl-lysine deacetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0048742 | regulation of skeletal muscle fiber development | IGI | J:127416 | |||||||||
| Biological Process | GO:0048742 | regulation of skeletal muscle fiber development | IGI | J:127416 | |||||||||
| Biological Process | GO:1902809 | regulation of skeletal muscle fiber differentiation | IGI | J:127416 | |||||||||
| Biological Process | GO:1902809 | regulation of skeletal muscle fiber differentiation | IGI | J:127416 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||