|
Symbol Name ID |
Inava
innate immunity activator MGI:1921579 |
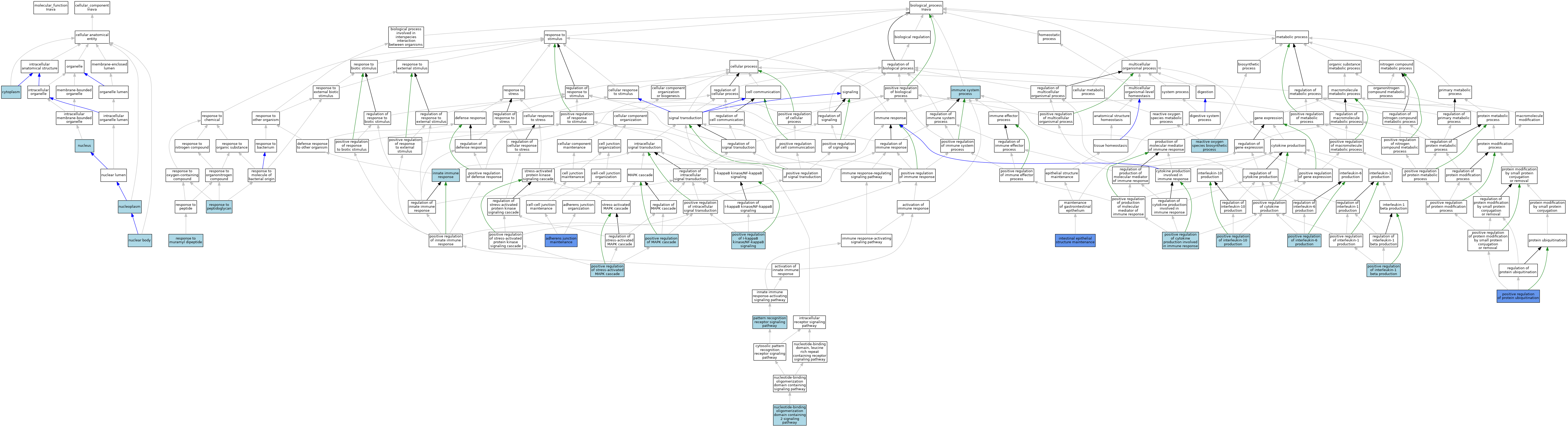
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0034334 | adherens junction maintenance | IDA | J:257317 | |||||||||
| Biological Process | GO:0034334 | adherens junction maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:0034334 | adherens junction maintenance | IBA | J:265628 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0060729 | intestinal epithelial structure maintenance | IMP | J:257317 | |||||||||
| Biological Process | GO:0070431 | nucleotide-binding oligomerization domain containing 2 signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0002221 | pattern recognition receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0002720 | positive regulation of cytokine production involved in immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB signaling | ISO | J:164563 | |||||||||
| Biological Process | GO:0032731 | positive regulation of interleukin-1 beta production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032733 | positive regulation of interleukin-10 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0032755 | positive regulation of interleukin-6 production | ISO | J:164563 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | IMP | J:257317 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0031398 | positive regulation of protein ubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0032874 | positive regulation of stress-activated MAPK cascade | ISO | J:164563 | |||||||||
| Biological Process | GO:1903409 | reactive oxygen species biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0032495 | response to muramyl dipeptide | ISO | J:164563 | |||||||||
| Biological Process | GO:0032494 | response to peptidoglycan | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||