|
Symbol Name ID |
Rpa1
replication protein A1 MGI:1915525 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
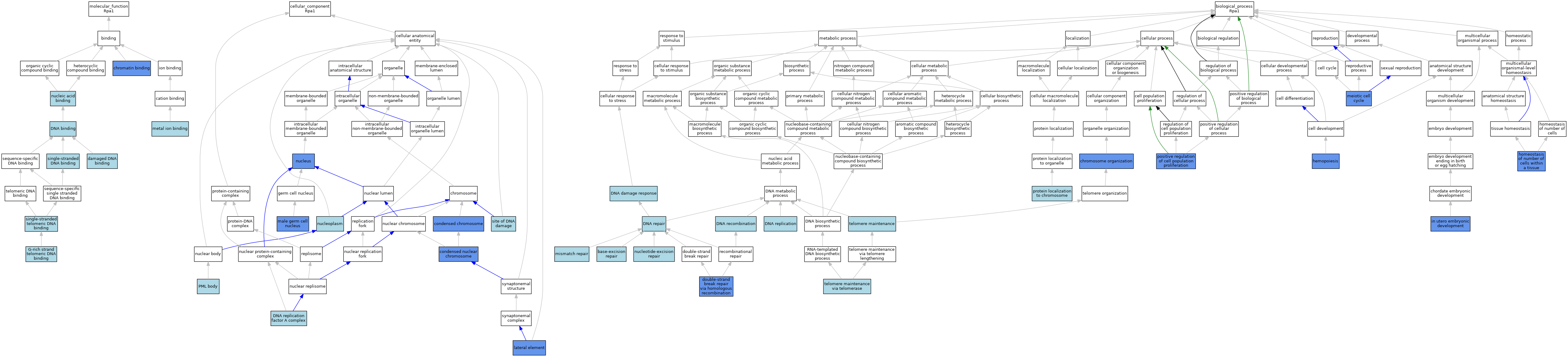
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:78200 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0098505 | G-rich strand telomeric DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0043047 | single-stranded telomeric DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000793 | condensed chromosome | IDA | J:78200 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:161203 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:182264 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:135578 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:131722 | |||||||||
| Cellular Component | GO:0005662 | DNA replication factor A complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005662 | DNA replication factor A complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005662 | DNA replication factor A complex | ISO | J:73065 | |||||||||
| Cellular Component | GO:0000800 | lateral element | IDA | J:176168 | |||||||||
| Cellular Component | GO:0000800 | lateral element | IDA | J:185266 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:123677 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912421 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912431 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912462 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912487 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:71817 | |||||||||
| Cellular Component | GO:0016605 | PML body | ISO | J:73065 | |||||||||
| Cellular Component | GO:0090734 | site of DNA damage | ISO | J:164563 | |||||||||
| Biological Process | GO:0006284 | base-excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0051276 | chromosome organization | IMP | J:99303 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | ISO | J:164563 | |||||||||
| Biological Process | GO:0006310 | DNA recombination | IEA | J:60000 | |||||||||
| Biological Process | GO:0006310 | DNA recombination | IEA | J:72247 | |||||||||
| Biological Process | GO:0006281 | DNA repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006260 | DNA replication | ISO | J:164563 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IMP | J:93303 | |||||||||
| Biological Process | GO:0030097 | hemopoiesis | IMP | J:99303 | |||||||||
| Biological Process | GO:0048873 | homeostasis of number of cells within a tissue | IMP | J:99303 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IMP | J:93303 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IBA | J:265628 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IDA | J:78200 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IBA | J:265628 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IMP | J:99303 | |||||||||
| Biological Process | GO:0034502 | protein localization to chromosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0000723 | telomere maintenance | ISO | J:164563 | |||||||||
| Biological Process | GO:0007004 | telomere maintenance via telomerase | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||