|
Symbol Name ID |
Arhgef5
Rho guanine nucleotide exchange factor 5 MGI:1858952 |
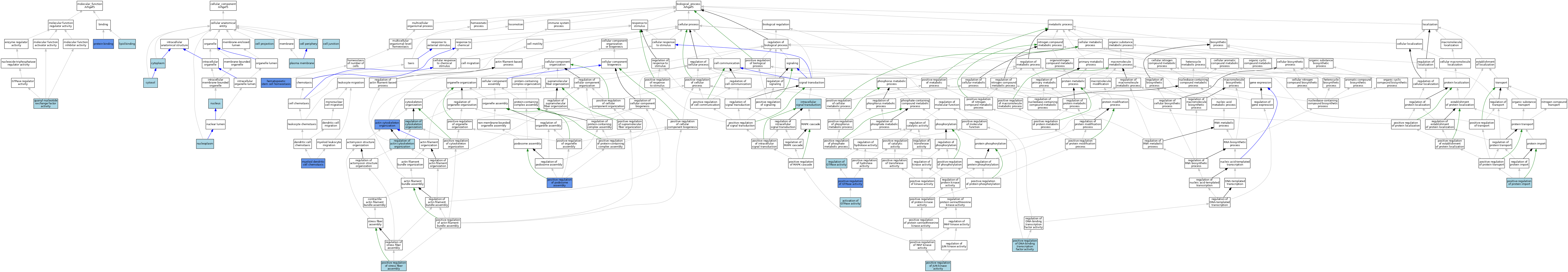
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182942 | |||||||||
| Cellular Component | GO:0030054 | cell junction | IEA | J:60000 | |||||||||
| Cellular Component | GO:0071944 | cell periphery | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0030036 | actin cytoskeleton organization | IDA | J:182942 | |||||||||
| Biological Process | GO:0090630 | activation of GTPase activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0061484 | hematopoietic stem cell homeostasis | IMP | J:232932 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | IEA | J:72247 | |||||||||
| Biological Process | GO:0002408 | myeloid dendritic cell chemotaxis | IMP | J:157812 | |||||||||
| Biological Process | GO:0051091 | positive regulation of DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0043547 | positive regulation of GTPase activity | IDA | J:182942 | |||||||||
| Biological Process | GO:0043507 | positive regulation of JUN kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0071803 | positive regulation of podosome assembly | IGI | J:182942 | |||||||||
| Biological Process | GO:0071803 | positive regulation of podosome assembly | IGI | J:182942 | |||||||||
| Biological Process | GO:1904591 | positive regulation of protein import | ISO | J:164563 | |||||||||
| Biological Process | GO:0051496 | positive regulation of stress fiber assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0032956 | regulation of actin cytoskeleton organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0051493 | regulation of cytoskeleton organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0043087 | regulation of GTPase activity | ISO | J:73065 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||