|
Symbol Name ID |
Exo1
exonuclease 1 MGI:1349427 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
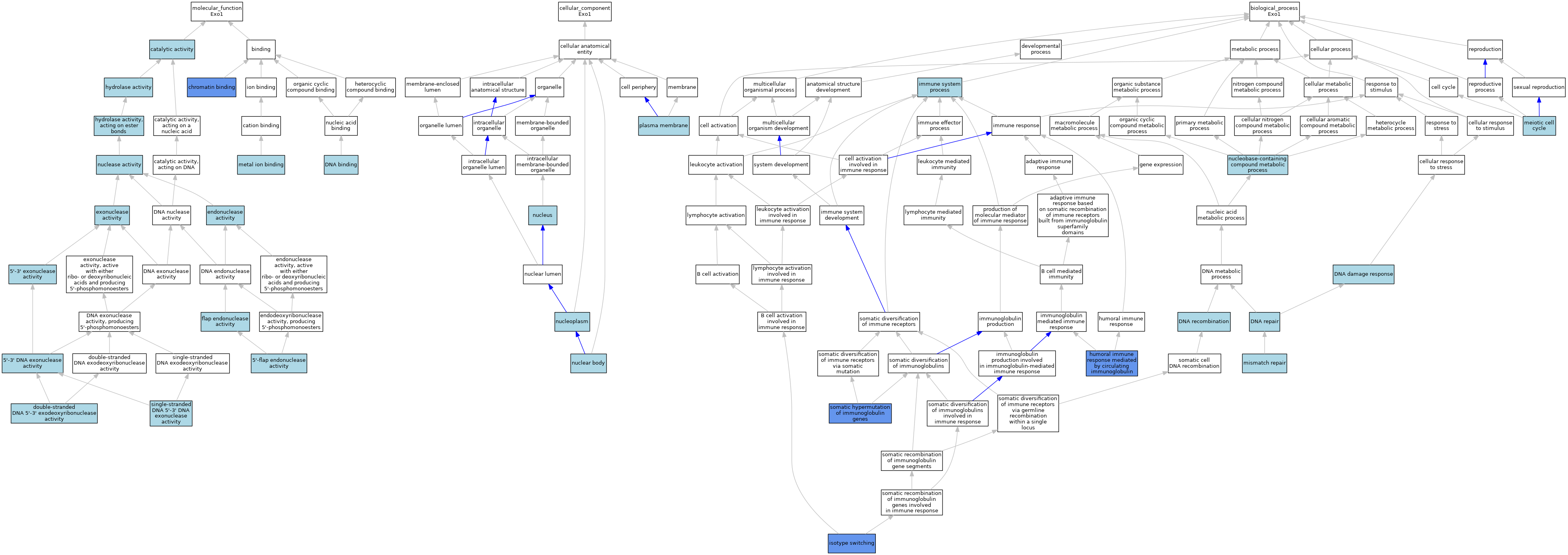
| Molecular Function | GO:0035312 | 5'-3' DNA exonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035312 | 5'-3' DNA exonuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008409 | 5'-3' exonuclease activity | ISO | J:58084 | |||||||||
| Molecular Function | GO:0017108 | 5'-flap endonuclease activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003824 | catalytic activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:87869 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004519 | endonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004527 | exonuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0048256 | flap endonuclease activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016788 | hydrolase activity, acting on ester bonds | IEA | J:72247 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004518 | nuclease activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0045145 | single-stranded DNA 5'-3' DNA exonuclease activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006310 | DNA recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0006310 | DNA recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:72247 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:60000 | |||||||||
| Biological Process | GO:0002455 | humoral immune response mediated by circulating immunoglobulin | IMP | J:87869 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045190 | isotype switching | IMP | J:87869 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | IBA | J:265628 | |||||||||
| Biological Process | GO:0006139 | nucleobase-containing compound metabolic process | ISO | J:58084 | |||||||||
| Biological Process | GO:0016446 | somatic hypermutation of immunoglobulin genes | IMP | J:87869 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||