|
Symbol Name ID |
Sort1
sortilin 1 MGI:1338015 |
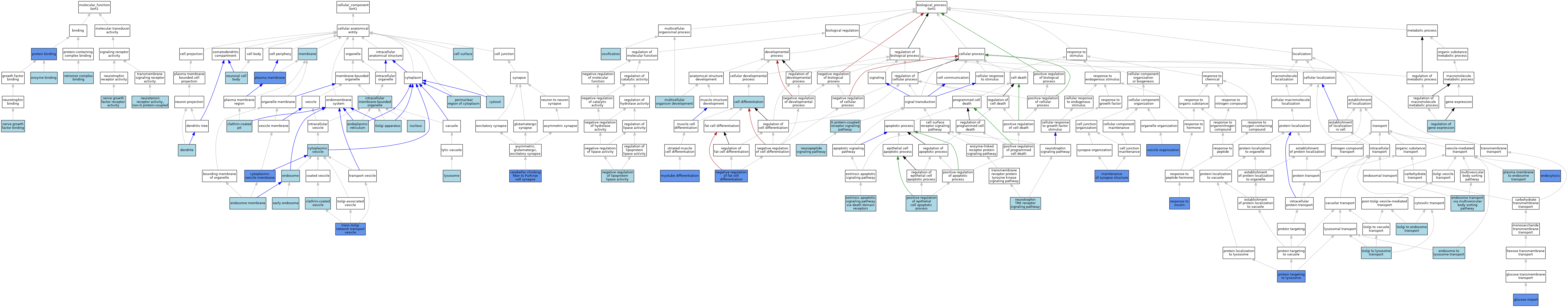
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0048406 | nerve growth factor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0010465 | nerve growth factor receptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0030379 | neurotensin receptor activity, non-G protein-coupled | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:136228 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:193410 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:159842 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:167062 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:208015 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:170255 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:149884 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:216244 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:216244 | |||||||||
| Molecular Function | GO:1905394 | retromer complex binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0150053 | cerebellar climbing fiber to Purkinje cell synapse | IDA | J:261277 | |||||||||
| Cellular Component | GO:0150053 | cerebellar climbing fiber to Purkinje cell synapse | IMP | J:261277 | |||||||||
| Cellular Component | GO:0150053 | cerebellar climbing fiber to Purkinje cell synapse | IDA | J:261277 | |||||||||
| Cellular Component | GO:0150053 | cerebellar climbing fiber to Purkinje cell synapse | IDA | J:261277 | |||||||||
| Cellular Component | GO:0005905 | clathrin-coated pit | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030136 | clathrin-coated vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030659 | cytoplasmic vesicle membrane | IDA | J:80816 | |||||||||
| Cellular Component | GO:0030659 | cytoplasmic vesicle membrane | IDA | J:234039 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0010008 | endosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005794 | Golgi apparatus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005764 | lysosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IEA | J:60000 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:99400 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030140 | trans-Golgi network transport vesicle | IDA | J:89488 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0006897 | endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0006897 | endocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0006897 | endocytosis | IMP | J:206213 | |||||||||
| Biological Process | GO:0008333 | endosome to lysosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0032509 | endosome transport via multivesicular body sorting pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors | ISO | J:164563 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0046323 | glucose import | IMP | J:99400 | |||||||||
| Biological Process | GO:0046323 | glucose import | ISO | J:164563 | |||||||||
| Biological Process | GO:0006895 | Golgi to endosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0006895 | Golgi to endosome transport | IBA | J:265628 | |||||||||
| Biological Process | GO:0090160 | Golgi to lysosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:0099558 | maintenance of synapse structure | IMP | J:261277 | |||||||||
| Biological Process | GO:0099558 | maintenance of synapse structure | IDA | J:261277 | |||||||||
| Biological Process | GO:0099558 | maintenance of synapse structure | IDA | J:261277 | |||||||||
| Biological Process | GO:0099558 | maintenance of synapse structure | IDA | J:261277 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0014902 | myotube differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0014902 | myotube differentiation | IMP | J:133006 | |||||||||
| Biological Process | GO:0045599 | negative regulation of fat cell differentiation | IDA | J:216244 | |||||||||
| Biological Process | GO:0051005 | negative regulation of lipoprotein lipase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0007218 | neuropeptide signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0048011 | neurotrophin TRK receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0001503 | ossification | IEA | J:60000 | |||||||||
| Biological Process | GO:0048227 | plasma membrane to endosome transport | ISO | J:164563 | |||||||||
| Biological Process | GO:1904037 | positive regulation of epithelial cell apoptotic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006622 | protein targeting to lysosome | IMP | J:154468 | |||||||||
| Biological Process | GO:0010468 | regulation of gene expression | ISO | J:164563 | |||||||||
| Biological Process | GO:0032868 | response to insulin | IMP | J:99400 | |||||||||
| Biological Process | GO:0032868 | response to insulin | ISO | J:164563 | |||||||||
| Biological Process | GO:0016050 | vesicle organization | ISO | J:164563 | |||||||||
| Biological Process | GO:0016050 | vesicle organization | IMP | J:99400 | |||||||||
| Biological Process | GO:0016050 | vesicle organization | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||