|
Symbol Name ID |
Fem1b
fem 1 homolog b MGI:1335087 |
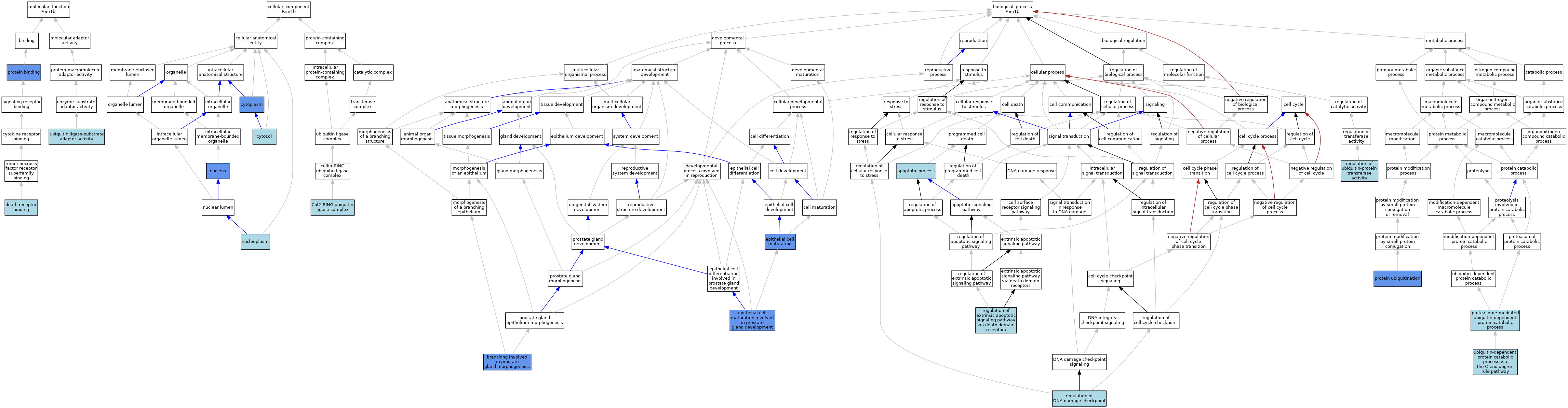
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005123 | death receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:139640 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:176478 | |||||||||
| Molecular Function | GO:1990756 | ubiquitin ligase-substrate adaptor activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031462 | Cul2-RING ubiquitin ligase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:176478 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:154112 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0060442 | branching involved in prostate gland morphogenesis | IMP | J:139640 | |||||||||
| Biological Process | GO:0002070 | epithelial cell maturation | IMP | J:139640 | |||||||||
| Biological Process | GO:0060743 | epithelial cell maturation involved in prostate gland development | IMP | J:139640 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | IDA | J:176478 | |||||||||
| Biological Process | GO:2000001 | regulation of DNA damage checkpoint | ISO | J:164563 | |||||||||
| Biological Process | GO:1902041 | regulation of extrinsic apoptotic signaling pathway via death domain receptors | ISO | J:164563 | |||||||||
| Biological Process | GO:0051438 | regulation of ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0140627 | ubiquitin-dependent protein catabolic process via the C-end degron rule pathway | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||