|
Symbol Name ID |
Ap2m1
adaptor-related protein complex 2, mu 1 subunit MGI:1298405 |
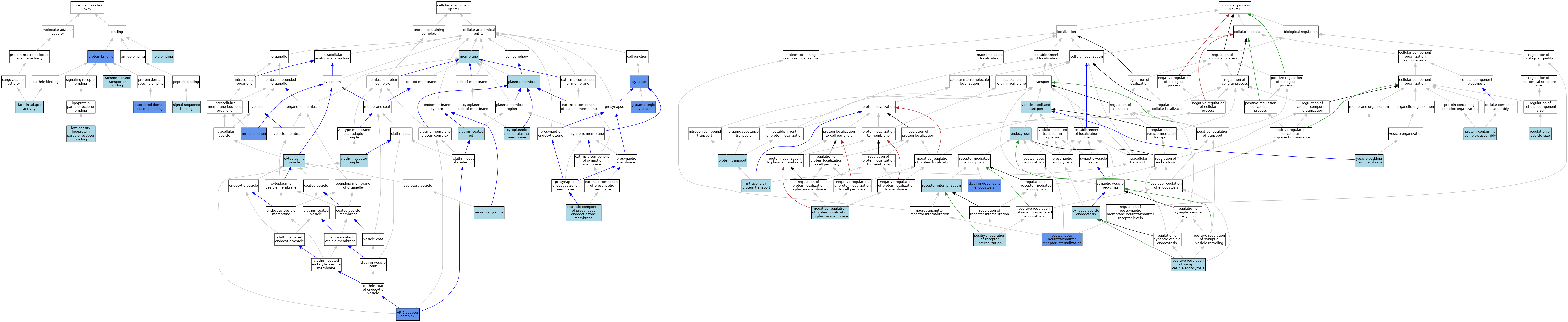
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0035615 | clathrin adaptor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0097718 | disordered domain specific binding | IDA | J:240276 | |||||||||
| Molecular Function | GO:0008289 | lipid binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0050750 | low-density lipoprotein particle receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:201438 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:240276 | |||||||||
| Molecular Function | GO:0005048 | signal sequence binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005048 | signal sequence binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0044325 | transmembrane transporter binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030122 | AP-2 adaptor complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030122 | AP-2 adaptor complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030122 | AP-2 adaptor complex | IDA | J:201438 | |||||||||
| Cellular Component | GO:0030122 | AP-2 adaptor complex | NAS | J:240303 | |||||||||
| Cellular Component | GO:0030122 | AP-2 adaptor complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030131 | clathrin adaptor complex | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005905 | clathrin-coated pit | ISO | J:164563 | |||||||||
| Cellular Component | GO:0009898 | cytoplasmic side of plasma membrane | NAS | J:320128 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IBA | J:265628 | |||||||||
| Cellular Component | GO:0098894 | extrinsic component of presynaptic endocytic zone membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:238446 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IDA | J:238446 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | IMP | J:238446 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030141 | secretory granule | TAS | J:48443 | |||||||||
| Cellular Component | GO:0045202 | synapse | IDA | J:263419 | |||||||||
| Cellular Component | GO:0045202 | synapse | EXP | J:263419 | |||||||||
| Biological Process | GO:0072583 | clathrin-dependent endocytosis | NAS | J:320128 | |||||||||
| Biological Process | GO:0072583 | clathrin-dependent endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0072583 | clathrin-dependent endocytosis | IDA | J:201438 | |||||||||
| Biological Process | GO:0072583 | clathrin-dependent endocytosis | IBA | J:265628 | |||||||||
| Biological Process | GO:0006897 | endocytosis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006886 | intracellular protein transport | TAS | J:48443 | |||||||||
| Biological Process | GO:1903077 | negative regulation of protein localization to plasma membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0002092 | positive regulation of receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:1900244 | positive regulation of synaptic vesicle endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0098884 | postsynaptic neurotransmitter receptor internalization | NAS | J:319684 | |||||||||
| Biological Process | GO:0098884 | postsynaptic neurotransmitter receptor internalization | ISO | J:155856 | |||||||||
| Biological Process | GO:0098884 | postsynaptic neurotransmitter receptor internalization | IMP | J:238446 | |||||||||
| Biological Process | GO:0098884 | postsynaptic neurotransmitter receptor internalization | IDA | J:238446 | |||||||||
| Biological Process | GO:0098884 | postsynaptic neurotransmitter receptor internalization | IDA | J:238446 | |||||||||
| Biological Process | GO:0015031 | protein transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0065003 | protein-containing complex assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0031623 | receptor internalization | ISO | J:164563 | |||||||||
| Biological Process | GO:0097494 | regulation of vesicle size | ISO | J:164563 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | ISO | J:164563 | |||||||||
| Biological Process | GO:0048488 | synaptic vesicle endocytosis | ISO | J:155856 | |||||||||
| Biological Process | GO:0006900 | vesicle budding from membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0016192 | vesicle-mediated transport | NAS | J:319992 | |||||||||
| Biological Process | GO:0016192 | vesicle-mediated transport | TAS | J:48443 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||