|
Symbol Name ID |
Irx3
Iroquois related homeobox 3 MGI:1197522 |
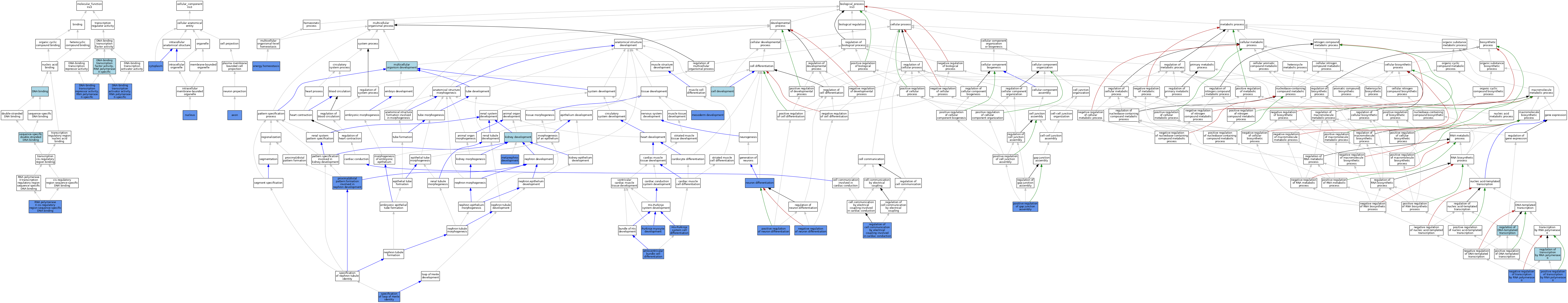
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | IDA | J:175992 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | IDA | J:175992 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IDA | J:175992 | |||||||||
| Molecular Function | GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:1990837 | sequence-specific double-stranded DNA binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0030424 | axon | IDA | J:152175 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:133640 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:133640 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:133640 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:133640 | |||||||||
| Biological Process | GO:0003167 | atrioventricular bundle cell differentiation | IMP | J:250159 | |||||||||
| Biological Process | GO:0048468 | cell development | IBA | J:265628 | |||||||||
| Biological Process | GO:0097009 | energy homeostasis | IDA | J:208887 | |||||||||
| Biological Process | GO:0060932 | His-Purkinje system cell differentiation | IMP | J:250159 | |||||||||
| Biological Process | GO:0001822 | kidney development | ISO | J:73065 | |||||||||
| Biological Process | GO:0007498 | mesoderm development | IGI | J:66591 | |||||||||
| Biological Process | GO:0001656 | metanephros development | IEP | J:125316 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0045665 | negative regulation of neuron differentiation | IDA | J:133641 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:175992 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IBA | J:265628 | |||||||||
| Biological Process | GO:0030182 | neuron differentiation | IDA | J:133641 | |||||||||
| Biological Process | GO:1903598 | positive regulation of gap junction assembly | IMP | J:250159 | |||||||||
| Biological Process | GO:1903598 | positive regulation of gap junction assembly | IMP | J:175992 | |||||||||
| Biological Process | GO:0045666 | positive regulation of neuron differentiation | IDA | J:133641 | |||||||||
| Biological Process | GO:0045944 | positive regulation of transcription by RNA polymerase II | IDA | J:175992 | |||||||||
| Biological Process | GO:0072047 | proximal/distal pattern formation involved in nephron development | IEP | J:125316 | |||||||||
| Biological Process | GO:0003165 | Purkinje myocyte development | IMP | J:250159 | |||||||||
| Biological Process | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction | IMP | J:175992 | |||||||||
| Biological Process | GO:0006355 | regulation of DNA-templated transcription | IEA | J:72247 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0072086 | specification of loop of Henle identity | IEP | J:125316 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||