|
Symbol Name ID |
Dmc1
DNA meiotic recombinase 1 MGI:105393 |
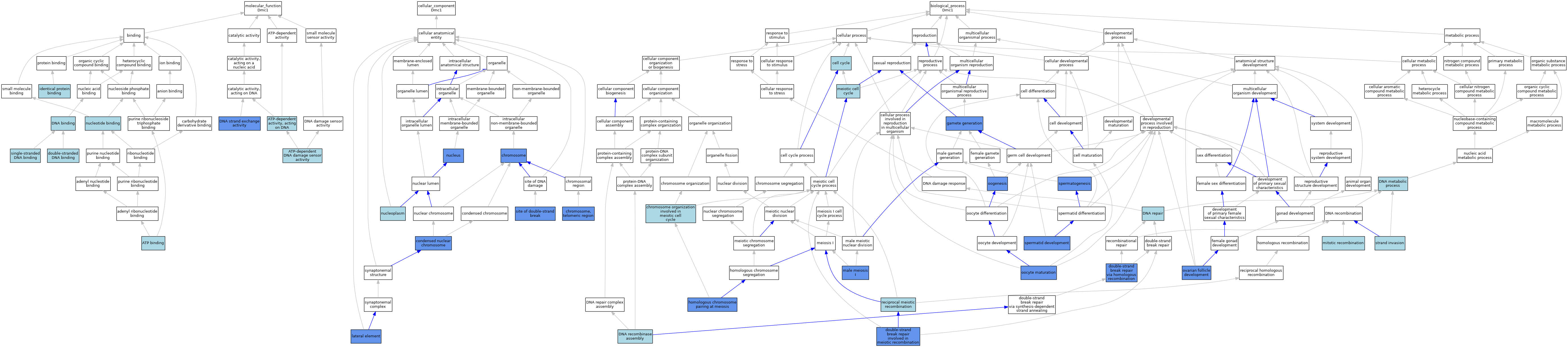
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0008094 | ATP-dependent activity, acting on DNA | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008094 | ATP-dependent activity, acting on DNA | IBA | J:265628 | |||||||||
| Molecular Function | GO:0140664 | ATP-dependent DNA damage sensor activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0000150 | DNA strand exchange activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000150 | DNA strand exchange activity | IDA | J:330108 | |||||||||
| Molecular Function | GO:0003690 | double-stranded DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:297328 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:291221 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:183982 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:285969 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:273173 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IDA | J:170340 | |||||||||
| Cellular Component | GO:0000781 | chromosome, telomeric region | IDA | J:158887 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:161203 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:236226 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:135578 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:213984 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:182264 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:131722 | |||||||||
| Cellular Component | GO:0000794 | condensed nuclear chromosome | IDA | J:185266 | |||||||||
| Cellular Component | GO:0000800 | lateral element | IDA | J:270644 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912421 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912431 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-912462 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:170340 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:185766 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | IDA | J:291221 | |||||||||
| Cellular Component | GO:0035861 | site of double-strand break | IDA | J:273173 | |||||||||
| Biological Process | GO:0007049 | cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0070192 | chromosome organization involved in meiotic cell cycle | IBA | J:265628 | |||||||||
| Biological Process | GO:0006259 | DNA metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0000730 | DNA recombinase assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IEA | J:72247 | |||||||||
| Biological Process | GO:1990918 | double-strand break repair involved in meiotic recombination | IDA | J:330108 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | IDA | J:330108 | |||||||||
| Biological Process | GO:0007276 | gamete generation | IMP | J:86161 | |||||||||
| Biological Process | GO:0007129 | homologous chromosome pairing at meiosis | IMP | J:123677 | |||||||||
| Biological Process | GO:0007141 | male meiosis I | IDA | J:330108 | |||||||||
| Biological Process | GO:0007141 | male meiosis I | IMP | J:123677 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0006312 | mitotic recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0001556 | oocyte maturation | IMP | J:123677 | |||||||||
| Biological Process | GO:0048477 | oogenesis | IGI | J:103876 | |||||||||
| Biological Process | GO:0048477 | oogenesis | IMP | J:103876 | |||||||||
| Biological Process | GO:0001541 | ovarian follicle development | IMP | J:96250 | |||||||||
| Biological Process | GO:0007131 | reciprocal meiotic recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0007131 | reciprocal meiotic recombination | IBA | J:265628 | |||||||||
| Biological Process | GO:0007286 | spermatid development | IMP | J:123677 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:86161 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:103876 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IGI | J:103876 | |||||||||
| Biological Process | GO:0042148 | strand invasion | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||