|
Symbol Name ID |
Xpc
xeroderma pigmentosum, complementation group C MGI:103557 |
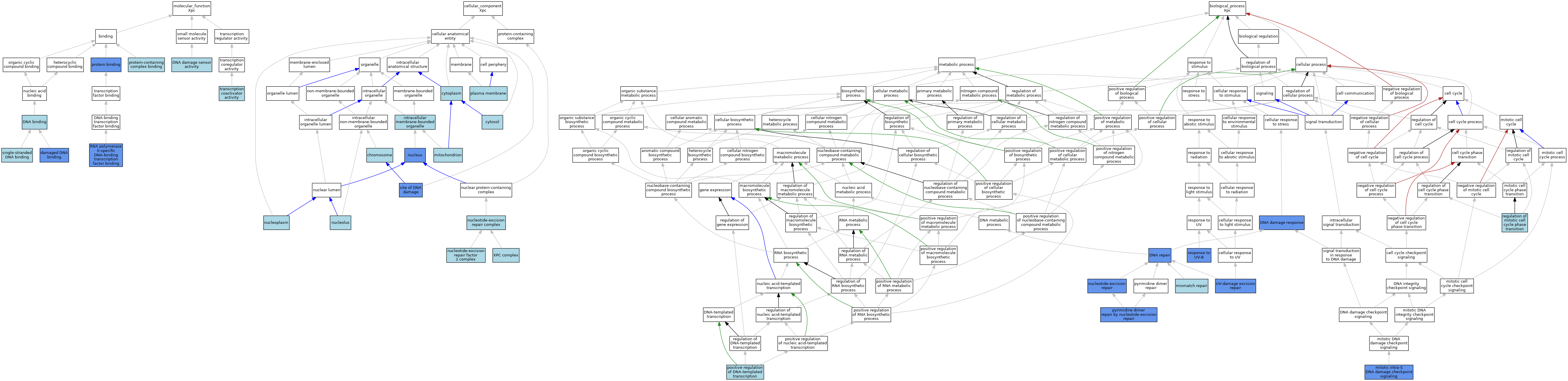
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003684 | damaged DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003684 | damaged DNA binding | IDA | J:185050 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0140612 | DNA damage sensor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:185050 | |||||||||
| Molecular Function | GO:0044877 | protein-containing complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | IDA | J:221330 | |||||||||
| Molecular Function | GO:0061629 | RNA polymerase II-specific DNA-binding transcription factor binding | IPI | J:221330 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003697 | single-stranded DNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003713 | transcription coactivator activity | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000109 | nucleotide-excision repair complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000111 | nucleotide-excision repair factor 2 complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:84299 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0090734 | site of DNA damage | IDA | J:185050 | |||||||||
| Cellular Component | GO:0071942 | XPC complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0071942 | XPC complex | IBA | J:265628 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IMP | J:88558 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IMP | J:80212 | |||||||||
| Biological Process | GO:0006298 | mismatch repair | IBA | J:265628 | |||||||||
| Biological Process | GO:0031573 | mitotic intra-S DNA damage checkpoint signaling | IGI | J:94447 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IBA | J:265628 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IDA | J:185050 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:117815 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:153645 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:153645 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:153645 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:153645 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IDA | J:84299 | |||||||||
| Biological Process | GO:0006289 | nucleotide-excision repair | IMP | J:38645 | |||||||||
| Biological Process | GO:0045893 | positive regulation of DNA-templated transcription | ISO | J:164563 | |||||||||
| Biological Process | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair | IMP | J:117815 | |||||||||
| Biological Process | GO:1901990 | regulation of mitotic cell cycle phase transition | ISO | J:164563 | |||||||||
| Biological Process | GO:0010224 | response to UV-B | IMP | J:88558 | |||||||||
| Biological Process | GO:0070914 | UV-damage excision repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0070914 | UV-damage excision repair | IDA | J:185050 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||