|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
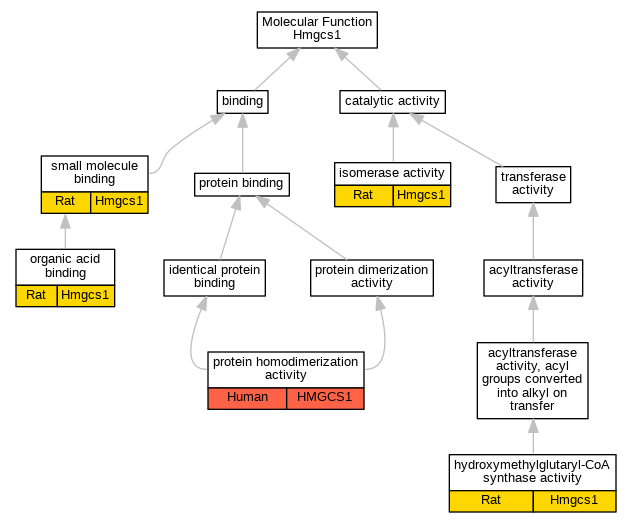
| Molecular Function | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity | Hmgcs1 | IDA | Rat | PMID:8094614|RGD:2326140 | |
| Molecular Function | GO:0016853 | isomerase activity | Hmgcs1 | IDA | Rat | PMID:17764185|RGD:2326141 | |
| Molecular Function | GO:0043177 | organic acid binding | Hmgcs1 | IDA | Rat | PMID:8094614|RGD:2326140 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | HMGCS1 | IDA | Human | PMID:20346956 | |
| Molecular Function | GO:0036094 | small molecule binding | Hmgcs1 | IDA | Rat | PMID:14984735|RGD:2316844 | |
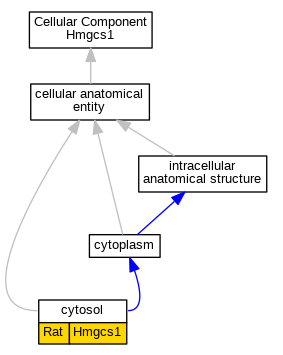
| Cellular Component | GO:0005829 | cytosol | Hmgcs1 | IDA | Rat | PMID:14984735|RGD:2316844 | |
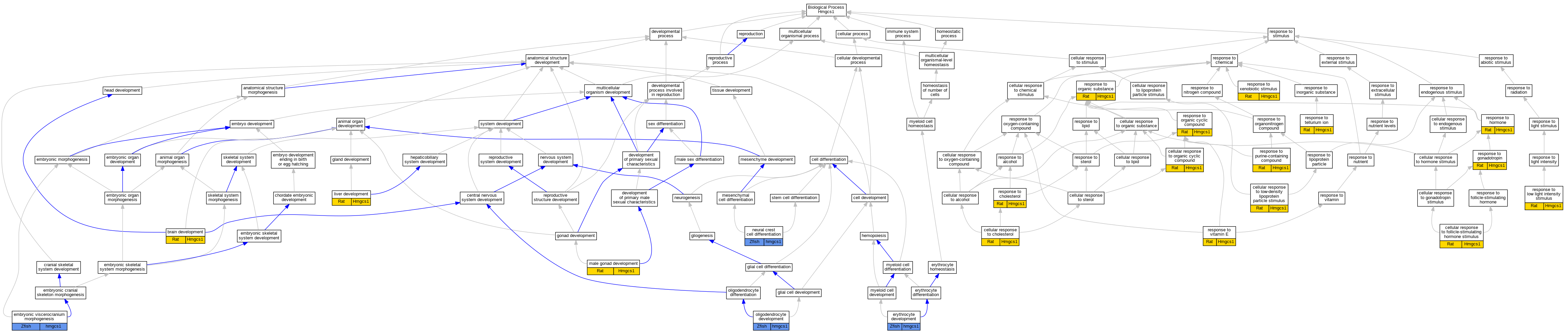
| Biological Process | GO:0007420 | brain development | Hmgcs1 | IEP | Rat | PMID:6133227|RGD:2326147 | |
| Biological Process | GO:0071397 | cellular response to cholesterol | Hmgcs1 | IEP | Rat | PMID:18303831|RGD:2326114 | |
| Biological Process | GO:0071372 | cellular response to follicle-stimulating hormone stimulus | Hmgcs1 | IEP | Rat | PMID:19196834|RGD:2326113 | |
| Biological Process | GO:0071404 | cellular response to low-density lipoprotein particle stimulus | Hmgcs1 | IDA | Rat | PMID:16260|RGD:2302235 | |
| Biological Process | GO:0071407 | cellular response to organic cyclic compound | Hmgcs1 | IEP | Rat | PMID:9409294|RGD:2326162 | |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | hmgcs1 | IMP | ZFIN:ZDB-GENO-210819-8 | Zfish | PMID:33197123 |
| Biological Process | GO:0048703 | embryonic viscerocranium morphogenesis | hmgcs1 | IMP | ZFIN:ZDB-GENO-170915-2 | Zfish | PMID:28686747 |
| Biological Process | GO:0048821 | erythrocyte development | hmgcs1 | IMP | ZFIN:ZDB-GENO-170915-2 | Zfish | PMID:30987969 |
| Biological Process | GO:0001889 | liver development | Hmgcs1 | IEP | Rat | PMID:5166591|RGD:2326169 | |
| Biological Process | GO:0008584 | male gonad development | Hmgcs1 | IEP | Rat | PMID:11792726|RGD:2326159 | |
| Biological Process | GO:0014033 | neural crest cell differentiation | hmgcs1 | IMP | ZFIN:ZDB-GENO-170915-2 | Zfish | PMID:28686747 |
| Biological Process | GO:0014003 | oligodendrocyte development | hmgcs1 | IMP | ZFIN:ZDB-GENO-140728-3 | Zfish | PMID:24573296 |
| Biological Process | GO:0070723 | response to cholesterol | Hmgcs1 | IEP | Rat | PMID:2903839|RGD:2326164 | |
| Biological Process | GO:0034698 | response to gonadotropin | Hmgcs1 | IEP | Rat | PMID:15459111|RGD:2326154 | |
| Biological Process | GO:0009725 | response to hormone | Hmgcs1 | IEP | Rat | PMID:8093833|RGD:2326142 | |
| Biological Process | GO:0009645 | response to low light intensity stimulus | Hmgcs1 | IEP | Rat | PMID:1683769|RGD:2326143 | |
| Biological Process | GO:0014070 | response to organic cyclic compound | Hmgcs1 | IEP | Rat | PMID:19527300|RGD:2326111 | |
| Biological Process | GO:0010033 | response to organic substance | Hmgcs1 | IEP | Rat | PMID:3670308|RGD:728568 | |
| Biological Process | GO:0014074 | response to purine-containing compound | Hmgcs1 | IDA | Rat | PMID:16260|RGD:2302235 | |
| Biological Process | GO:0046690 | response to tellurium ion | Hmgcs1 | IEP | Rat | PMID:10642751|RGD:2326161 | |
| Biological Process | GO:0033197 | response to vitamin E | Hmgcs1 | IEP | Rat | PMID:15753162|RGD:2326153 | |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Hmgcs1 | IEP | Rat | PMID:16020911|RGD:2326152 | |
| Biological Process | GO:0009410 | response to xenobiotic stimulus | Hmgcs1 | IEP | Rat | PMID:17311803|RGD:2302234 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||