|
Source |
Alliance of Genome Resources |
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
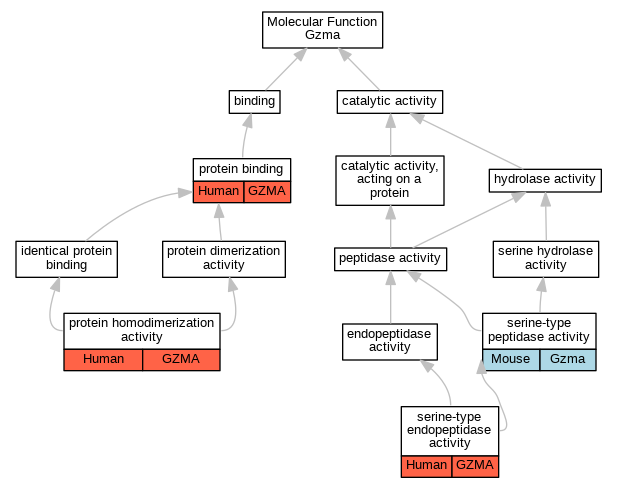
| Molecular Function | GO:0005515 | protein binding | GZMA | IPI | UniProtKB:P27695 | Human | PMID:12524539 |
| Molecular Function | GO:0042803 | protein homodimerization activity | GZMA | IDA | Human | PMID:12819770 | |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | GZMA | IDA | Human | PMID:11331782 | |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | GZMA | IDA | Human | PMID:12819770 | |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | GZMA | IDA | Human | PMID:32299851 | |
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | GZMA | IDA | Human | PMID:34022140 | |
| Molecular Function | GO:0008236 | serine-type peptidase activity | Gzma | IDA | Mouse | PMID:16618603 | |
| Molecular Function | GO:0008236 | serine-type peptidase activity | Gzma | IDA | Mouse | PMID:35482418 | |
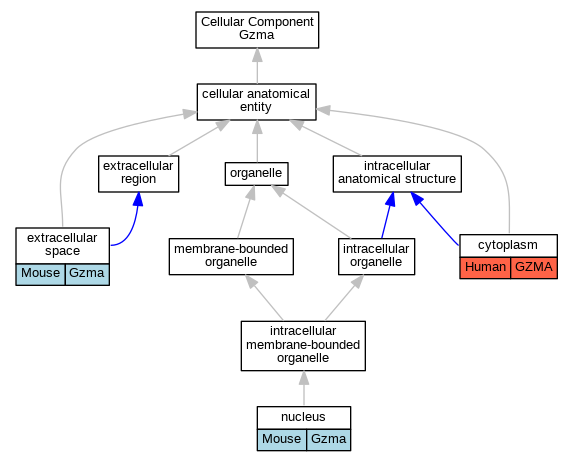
| Cellular Component | GO:0005737 | cytoplasm | GZMA | IDA | Human | PMID:20038786 | |
| Cellular Component | GO:0005737 | cytoplasm | GZMA | IDA | Human | PMID:34022140 | |
| Cellular Component | GO:0005615 | extracellular space | Gzma | IDA | Mouse | PMID:35482418 | |
| Cellular Component | GO:0005634 | nucleus | Gzma | IDA | Mouse | MGI:MGI:5546004|PMID:1860869 | |
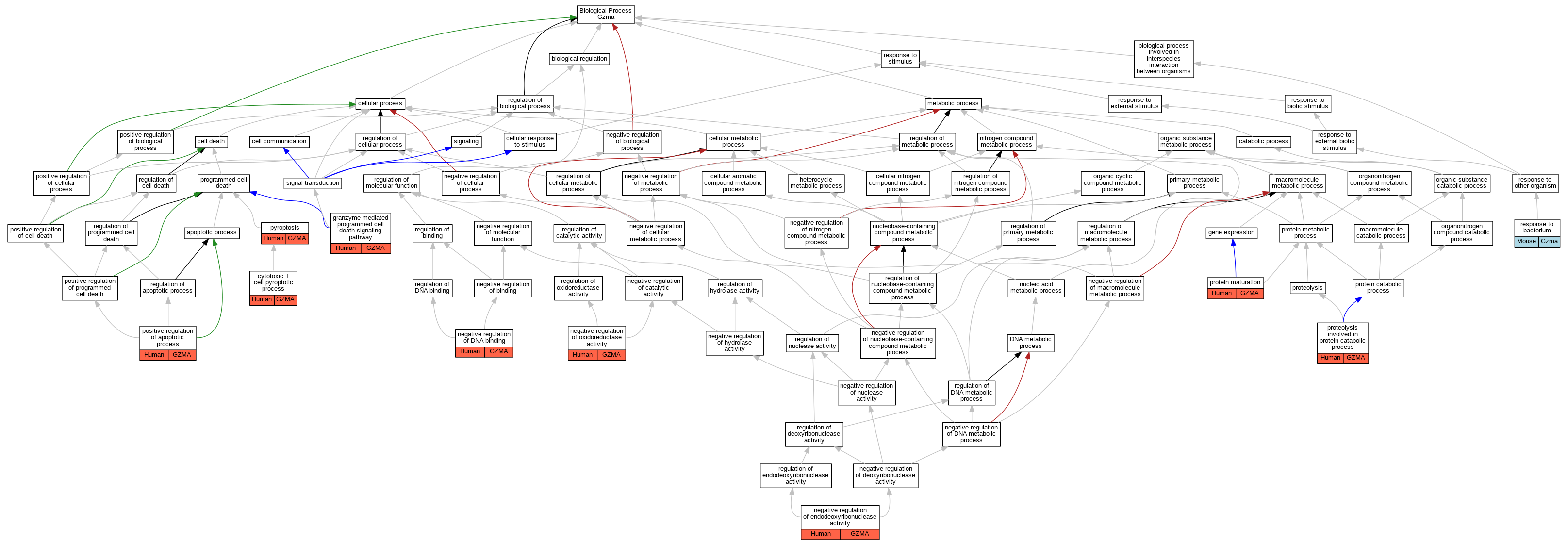
| Biological Process | GO:1902483 | cytotoxic T cell pyroptotic process | GZMA | IDA | Human | PMID:32299851 | |
| Biological Process | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | GZMA | IDA | Human | PMID:32299851 | |
| Biological Process | GO:0140507 | granzyme-mediated programmed cell death signaling pathway | GZMA | IDA | Human | PMID:34022140 | |
| Biological Process | GO:0043392 | negative regulation of DNA binding | GZMA | IDA | Human | PMID:12524539 | |
| Biological Process | GO:0032078 | negative regulation of endodeoxyribonuclease activity | GZMA | IDA | Human | PMID:12524539 | |
| Biological Process | GO:0051354 | negative regulation of oxidoreductase activity | GZMA | IDA | Human | PMID:12524539 | |
| Biological Process | GO:0043065 | positive regulation of apoptotic process | GZMA | IDA | Human | PMID:12524539 | |
| Biological Process | GO:0051604 | protein maturation | GZMA | IDA | Human | PMID:32299851 | |
| Biological Process | GO:0051604 | protein maturation | GZMA | IDA | Human | PMID:34022140 | |
| Biological Process | GO:0051603 | proteolysis involved in protein catabolic process | GZMA | IDA | Human | PMID:12524539 | |
| Biological Process | GO:0051603 | proteolysis involved in protein catabolic process | GZMA | IDA | Human | PMID:32299851 | |
| Biological Process | GO:0070269 | pyroptosis | GZMA | IDA | Human | PMID:32299851 | |
| Biological Process | GO:0009617 | response to bacterium | Gzma | IEP | Mouse | PMID:23012479 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||