Comparative GO Graph (mouse, human, rat, zebrafish)
Compare experimental GO annotations for Human-Mouse-Rat-Zebrafish orthologs to Mouse Scube2
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
- This class contains
- Human genes SCUBE2
- Mouse genes Scube2
- Rat genes Scube2
- Zfish genes scube2
- Annotations are indicated with nodes colored by organism:
Human,
Mouse,
Rat,
Zfish
- Relations between GO terms are indicated by colored edges "is_a"; "part_of"; "regulates"; "positively_regulates"; "negatively_regulates"
- GO annotations: Human from GOA; Mouse from MGI; Rat from RGD; Zebrafish from ZFIN.
- Only experimental annotations are displayed: EXP, IDA, IEP, IGI, IMP, IPI, HTP, HDA, HMP, HGI, HEP. Evidence codes are listed at the bottom of the page.
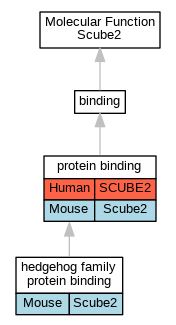
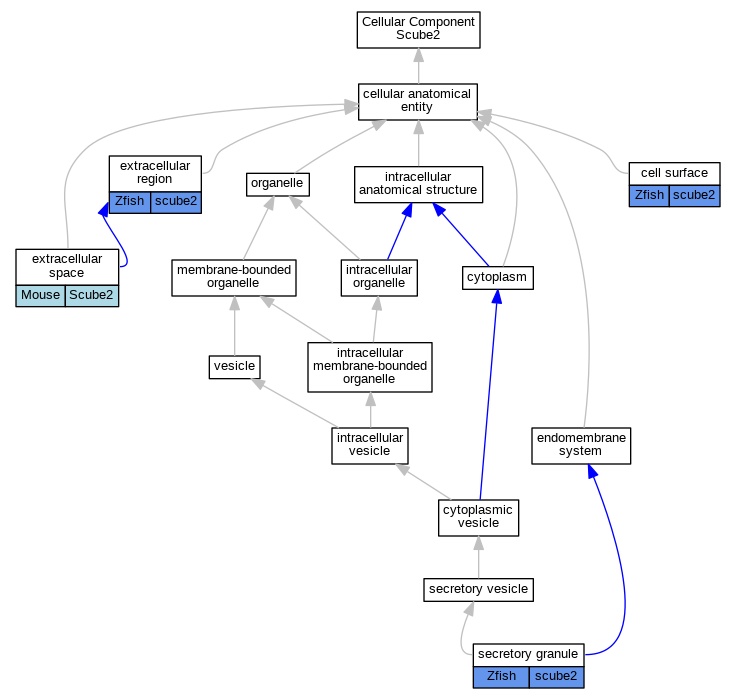
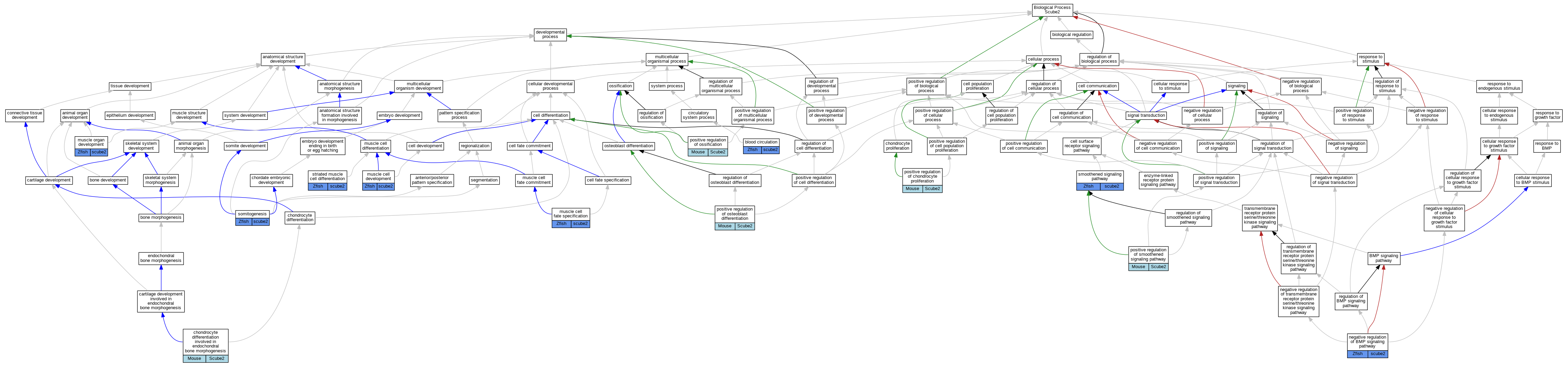 Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Scube2
Full set of experimental annotations for Human-Rat-Zebrafish genes considered orthologs of Mouse in the Alliance stringent orthology set Scube2
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0097108 | hedgehog family protein binding | Scube2 | IPI | PR:P97812 | Mouse | PMID:25639508 |
| Molecular Function | GO:0005515 | protein binding | SCUBE2 | IPI | UniProtKB:Q8IWY4 | Human | PMID:12270931 |
| Molecular Function | GO:0005515 | protein binding | Scube2 | IPI | PR:Q61115 | Mouse | PMID:25639508 |
| Cellular Component | GO:0009986 | cell surface | scube2 | IDA | | Zfish | PMID:15753045 |
| Cellular Component | GO:0005576 | extracellular region | scube2 | IDA | | Zfish | PMID:16626681 |
| Cellular Component | GO:0005615 | extracellular space | Scube2 | IDA | | Mouse | PMID:25639508 |
| Cellular Component | GO:0030141 | secretory granule | scube2 | IDA | | Zfish | PMID:15753045 |
| Biological Process | GO:0008015 | blood circulation | scube2 | IMP | ZFIN:ZDB-GENO-070808-35 | Zfish | PMID:9007249 |
| Biological Process | GO:0008015 | blood circulation | scube2 | IMP | ZFIN:ZDB-GENO-070808-24 | Zfish | PMID:9007249 |
| Biological Process | GO:0008015 | blood circulation | scube2 | IMP | ZFIN:ZDB-GENO-070808-38 | Zfish | PMID:9007249 |
| Biological Process | GO:0003413 | chondrocyte differentiation involved in endochondral bone morphogenesis | Scube2 | IMP | MGI:MGI:5698367 | Mouse | PMID:25639508 |
| Biological Process | GO:0055001 | muscle cell development | scube2 | IGI | ZFIN:ZDB-MRPHLNO-050510-1,ZFIN:ZDB-MRPHLNO-140114-6,ZFIN:ZDB-MRPHLNO-140114-7 | Zfish | PMID:24131633 |
| Biological Process | GO:0055001 | muscle cell development | scube2 | IGI | ZFIN:ZDB-MRPHLNO-050510-1,ZFIN:ZDB-MRPHLNO-120813-6,ZFIN:ZDB-MRPHLNO-120813-7 | Zfish | PMID:22609552 |
| Biological Process | GO:0042694 | muscle cell fate specification | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:10642786 |
| Biological Process | GO:0007517 | muscle organ development | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:15660164 |
| Biological Process | GO:0030514 | negative regulation of BMP signaling pathway | scube2 | IGI | | Zfish | PMID:15753045 |
| Biological Process | GO:1902732 | positive regulation of chondrocyte proliferation | Scube2 | IMP | MGI:MGI:5698367 | Mouse | PMID:25639508 |
| Biological Process | GO:0045778 | positive regulation of ossification | Scube2 | IMP | MGI:MGI:5698367 | Mouse | PMID:25639508 |
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | Scube2 | IGI | MGI:MGI:96533 | Mouse | PMID:25639508 |
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | Scube2 | IMP | | Mouse | PMID:25639508 |
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | Scube2 | IMP | MGI:MGI:5698367 | Mouse | PMID:25639508 |
| Biological Process | GO:0045669 | positive regulation of osteoblast differentiation | Scube2 | IDA | | Mouse | PMID:25639508 |
| Biological Process | GO:0045880 | positive regulation of smoothened signaling pathway | Scube2 | IDA | | Mouse | PMID:25639508 |
| Biological Process | GO:0007224 | smoothened signaling pathway | scube2 | IGI | ZFIN:ZDB-MRPHLNO-050510-1,ZFIN:ZDB-MRPHLNO-120813-6,ZFIN:ZDB-MRPHLNO-120813-7 | Zfish | PMID:22609552 |
| Biological Process | GO:0007224 | smoothened signaling pathway | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:16626681 |
| Biological Process | GO:0007224 | smoothened signaling pathway | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:15660164 |
| Biological Process | GO:0007224 | smoothened signaling pathway | scube2 | IGI | ZFIN:ZDB-GENO-050209-30,ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:16626681 |
| Biological Process | GO:0007224 | smoothened signaling pathway | scube2 | IGI | ZFIN:ZDB-GENE-980526-166,ZFIN:ZDB-MRPHLNO-050510-1,ZFIN:ZDB-MRPHLNO-120813-6,ZFIN:ZDB-MRPHLNO-120813-7 | Zfish | PMID:22609552 |
| Biological Process | GO:0001756 | somitogenesis | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:16626681 |
| Biological Process | GO:0001756 | somitogenesis | scube2 | IMP | ZFIN:ZDB-GENO-980202-1089,ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:16626681 |
| Biological Process | GO:0001756 | somitogenesis | scube2 | IMP | ZFIN:ZDB-GENO-980202-1089 | Zfish | PMID:16626681 |
| Biological Process | GO:0051146 | striated muscle cell differentiation | scube2 | IMP | ZFIN:ZDB-GENO-980202-1517 | Zfish | PMID:16626681 |
Gene Ontology Evidence Code Abbreviations:
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools





