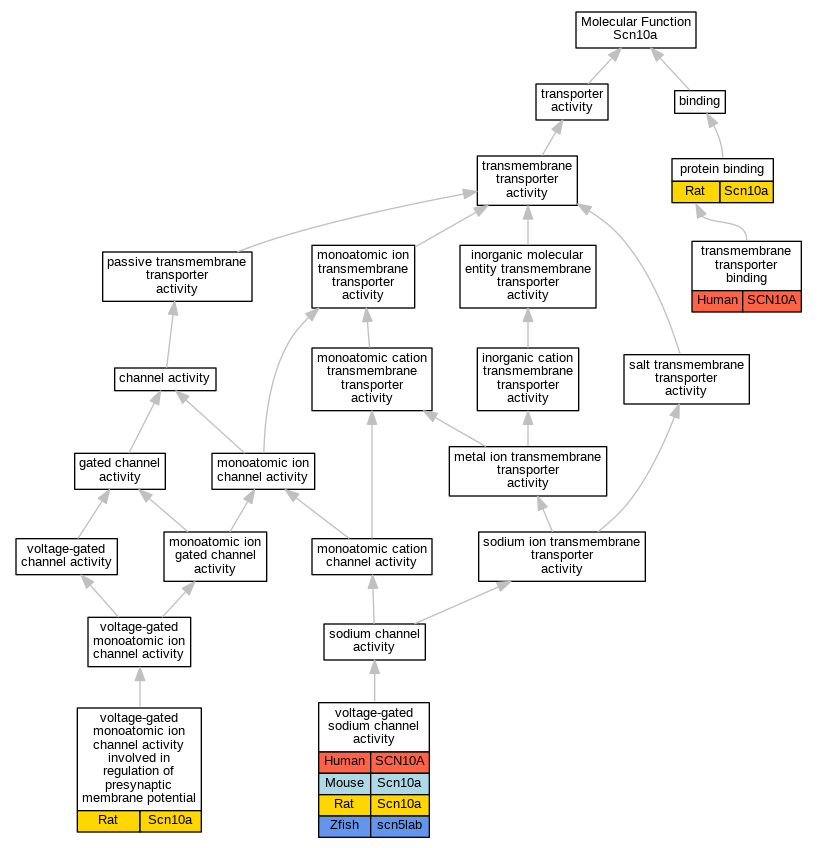
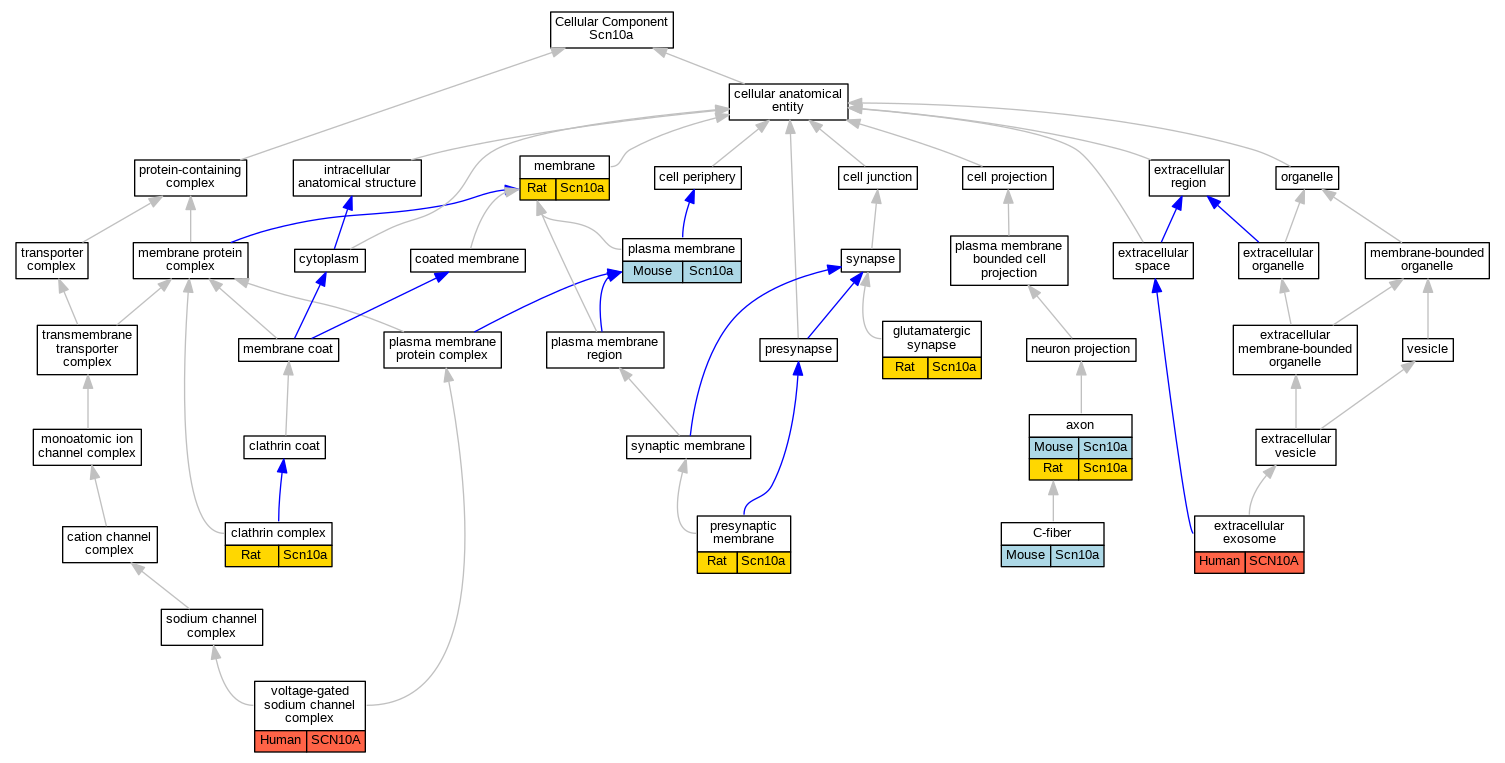
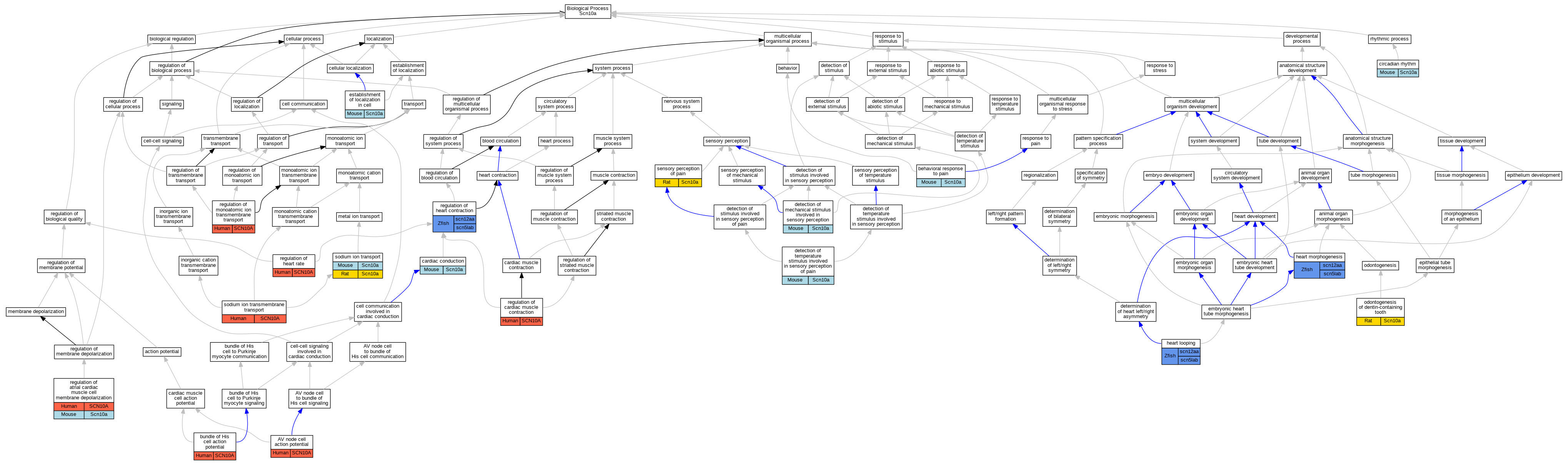
Graphs display curated GO classifications for mouse, human, rat and zebrafish orthologs annotated from the biomedical literature.
| Category |
ID |
Classification term |
Gene |
Evidence |
Inferred from |
Organism |
Reference |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | UniProtKB:P05943 | Rat | PMID:12050667|RGD:8554600 |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | UniProtKB:P05943 | Rat | PMID:12591166|RGD:8553968 |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | UniProtKB:Q63425 | Rat | PMID:12591166|RGD:8553968 |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | UniProtKB:Q9QZR8 | Rat | PMID:12591166|RGD:8553968 |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | UniProtKB:Q9Z336 | Rat | PMID:12591166|RGD:8553968 |
| Molecular Function | GO:0005515 | protein binding | Scn10a | IPI | RGD:628721 | Rat | PMID:15797711|RGD:1359053 |
| Molecular Function | GO:0044325 | transmembrane transporter binding | SCN10A | IPI | UniProtKB:Q14524 | Human | PMID:24998131 |
| Molecular Function | GO:0099508 | voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential | Scn10a | IDA | | Rat | PMID:26187311|RGD:155230761 |
| Molecular Function | GO:0099508 | voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential | Scn10a | IMP | | Rat | PMID:26187311|RGD:155230761 |
| Molecular Function | GO:0005248 | voltage-gated sodium channel activity | SCN10A | IDA | | Human | PMID:9839820 |
| Molecular Function | GO:0005248 | voltage-gated sodium channel activity | Scn10a | IDA | | Mouse | PMID:16029194 |
| Molecular Function | GO:0005248 | voltage-gated sodium channel activity | Scn10a | IDA | | Rat | PMID:8538791|RGD:69979 |
| Molecular Function | GO:0005248 | voltage-gated sodium channel activity | scn5lab | IDA | | Zfish | PMID:17623065 |
| Cellular Component | GO:0030424 | axon | Scn10a | IDA | | Mouse | PMID:16029194 |
| Cellular Component | GO:0030424 | axon | Scn10a | IDA | | Rat | PMID:21118538|RGD:15090835 |
| Cellular Component | GO:0044299 | C-fiber | Scn10a | IDA | | Mouse | PMID:16029194 |
| Cellular Component | GO:0071439 | clathrin complex | Scn10a | IDA | | Rat | PMID:15797711|RGD:1359053 |
| Cellular Component | GO:0070062 | extracellular exosome | SCN10A | HDA | | Human | PMID:19056867 |
| Cellular Component | GO:0098978 | glutamatergic synapse | Scn10a | IDA | | Rat | PMID:19162133|RGD:13702365 |
| Cellular Component | GO:0016020 | membrane | Scn10a | IDA | | Rat | PMID:8538791|RGD:69979 |
| Cellular Component | GO:0005886 | plasma membrane | Scn10a | IDA | | Mouse | PMID:16029194 |
| Cellular Component | GO:0042734 | presynaptic membrane | Scn10a | IDA | | Rat | PMID:19162133|RGD:13702365 |
| Cellular Component | GO:0001518 | voltage-gated sodium channel complex | SCN10A | IDA | | Human | PMID:9839820 |
| Biological Process | GO:0086016 | AV node cell action potential | SCN10A | IMP | | Human | PMID:24998131 |
| Biological Process | GO:0048266 | behavioral response to pain | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:24440715 |
| Biological Process | GO:0086043 | bundle of His cell action potential | SCN10A | IMP | | Human | PMID:24998131 |
| Biological Process | GO:0061337 | cardiac conduction | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:20062061 |
| Biological Process | GO:0007623 | circadian rhythm | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:25101983 |
| Biological Process | GO:0050974 | detection of mechanical stimulus involved in sensory perception | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:25101983 |
| Biological Process | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | Scn10a | IMP | MGI:MGI:4430300 | Mouse | PMID:19931571 |
| Biological Process | GO:0050965 | detection of temperature stimulus involved in sensory perception of pain | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:25101983 |
| Biological Process | GO:0051649 | establishment of localization in cell | Scn10a | IDA | | Mouse | PMID:16029194 |
| Biological Process | GO:0001947 | heart looping | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-2 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0001947 | heart looping | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-2 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0003007 | heart morphogenesis | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0042475 | odontogenesis of dentin-containing tooth | Scn10a | IEP | | Rat | PMID:21640979|RGD:6484223 |
| Biological Process | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | SCN10A | IMP | | Human | PMID:20062061 |
| Biological Process | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | SCN10A | IMP | | Human | PMID:21041692 |
| Biological Process | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization | Scn10a | IMP | MGI:MGI:2180183 | Mouse | PMID:20062061 |
| Biological Process | GO:0055117 | regulation of cardiac muscle contraction | SCN10A | IMP | | Human | PMID:20062061 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:25282101 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-2 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn12aa | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn12aa | IMP | ZFIN:ZDB-MRPHLNO-100616-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:25282101 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn5lab | IMP | ZFIN:ZDB-MRPHLNO-120105-1 | Zfish | PMID:20339120 |
| Biological Process | GO:0008016 | regulation of heart contraction | scn5lab | IGI | ZFIN:ZDB-MRPHLNO-100616-1,ZFIN:ZDB-MRPHLNO-100616-3 | Zfish | PMID:20339120 |
| Biological Process | GO:0002027 | regulation of heart rate | SCN10A | IMP | | Human | PMID:20062061 |
| Biological Process | GO:0002027 | regulation of heart rate | SCN10A | IMP | | Human | PMID:21041692 |
| Biological Process | GO:0034765 | regulation of monoatomic ion transmembrane transport | SCN10A | IDA | | Human | PMID:9839820 |
| Biological Process | GO:0019233 | sensory perception of pain | Scn10a | IMP | | Rat | PMID:21965668|RGD:6484251 |
| Biological Process | GO:0035725 | sodium ion transmembrane transport | SCN10A | IDA | | Human | PMID:9839820 |
| Biological Process | GO:0006814 | sodium ion transport | Scn10a | IDA | | Mouse | PMID:16029194 |
| Biological Process | GO:0006814 | sodium ion transport | Scn10a | IDA | | Rat | PMID:8538791|RGD:69979 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
 Analysis Tools
Analysis Tools