|
Source |
Alliance of Genome Resources |


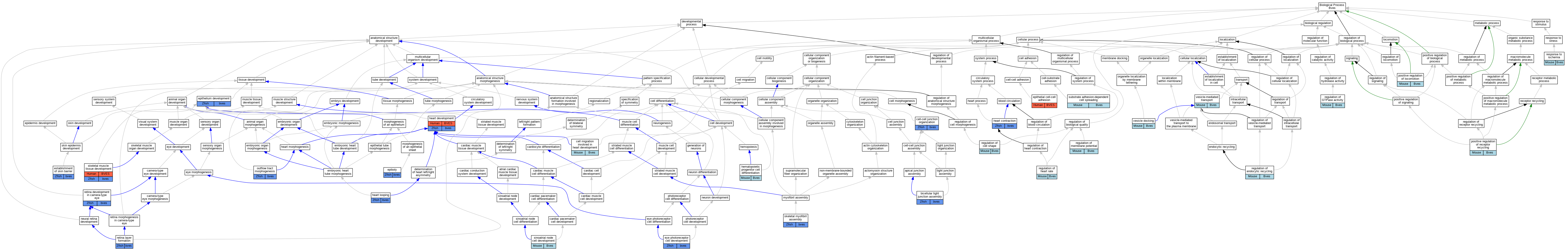
| Category | ID | Classification term | Gene | Evidence | Inferred from | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030552 | cAMP binding | Bves | IDA | Mouse | PMID:22354168 | |
| Molecular Function | GO:0005515 | protein binding | BVES | IPI | UniProtKB:P97438 | Human | PMID:26642364 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:P63024|UniProtKB:P63044 | Mouse | MGI:MGI:4431297|PMID:20057356 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:Q9CWR0 | Mouse | MGI:MGI:4457243|PMID:18541910 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:P39447 | Mouse | MGI:MGI:3607041|PMID:16188940 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:P51637 | Mouse | MGI:MGI:5550350|PMID:24066022 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:Q8BTG7 | Mouse | MGI:MGI:5613454|PMID:24048452 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | UniProtKB:P63025 | Mouse | MGI:MGI:4431297|PMID:20057356 |
| Molecular Function | GO:0005515 | protein binding | Bves | IPI | PR:P97438 | Mouse | PMID:22354168 |
| Molecular Function | GO:0005198 | structural molecule activity | BVES | IDA | Human | PMID:16188940 | |
| Cellular Component | GO:0005923 | bicellular tight junction | BVES | IDA | Human | PMID:16188940 | |
| Cellular Component | GO:0005923 | bicellular tight junction | Bves | IDA | Mouse | MGI:MGI:3607041|PMID:16188940 | |
| Cellular Component | GO:0005901 | caveola | Bves | IDA | Mouse | MGI:MGI:5550350|PMID:24066022 | |
| Cellular Component | GO:0030054 | cell junction | BVES | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0031253 | cell projection membrane | Bves | IDA | Mouse | MGI:MGI:5613454|PMID:24048452 | |
| Cellular Component | GO:0016328 | lateral plasma membrane | BVES | IDA | Human | PMID:16188940 | |
| Cellular Component | GO:0016328 | lateral plasma membrane | Bves | IDA | Mouse | MGI:MGI:3607041|PMID:16188940 | |
| Cellular Component | GO:0016020 | membrane | BVES | IDA | Human | PMID:16188940 | |
| Cellular Component | GO:0016020 | membrane | Bves | IDA | Mouse | MGI:MGI:3607041|PMID:16188940 | |
| Cellular Component | GO:0016020 | membrane | Bves | IDA | Mouse | PMID:10882522 | |
| Cellular Component | GO:0005886 | plasma membrane | BVES | IDA | Human | GO_REF:0000052 | |
| Cellular Component | GO:0005886 | plasma membrane | Bves | IDA | Mouse | MGI:MGI:4457243|PMID:18541910 | |
| Cellular Component | GO:0005886 | plasma membrane | Bves | IDA | Mouse | MGI:MGI:4431297|PMID:20057356 | |
| Cellular Component | GO:0005886 | plasma membrane | Bves | IDA | Mouse | PMID:22354168 | |
| Cellular Component | GO:0042383 | sarcolemma | BVES | IDA | Human | PMID:26642364 | |
| Cellular Component | GO:0042383 | sarcolemma | Bves | IDA | Mouse | MGI:MGI:5550350|PMID:24066022 | |
| Cellular Component | GO:0042383 | sarcolemma | bves | IDA | Zfish | PMID:26642364|ZFIN:ZDB-PUB-151208-1 | |
| Biological Process | GO:0070830 | bicellular tight junction assembly | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0070830 | bicellular tight junction assembly | bves | IMP | ZFIN:ZDB-MRPHLNO-130116-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0060973 | cell migration involved in heart development | Bves | IMP | Mouse | MGI:MGI:5613454|PMID:24048452 | |
| Biological Process | GO:0045216 | cell-cell junction organization | bves | IMP | Zfish | PMID:24741362 | |
| Biological Process | GO:0090504 | epiboly | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0090504 | epiboly | bves | IMP | ZFIN:ZDB-MRPHLNO-130116-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0090136 | epithelial cell-cell adhesion | BVES | IDA | Human | PMID:16188940 | |
| Biological Process | GO:0060429 | epithelium development | bves | IMP | ZFIN:ZDB-MRPHLNO-130116-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0060429 | epithelium development | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0061436 | establishment of skin barrier | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0061436 | establishment of skin barrier | bves | IMP | ZFIN:ZDB-MRPHLNO-130116-1 | Zfish | PMID:23019331 |
| Biological Process | GO:0042462 | eye photoreceptor cell development | bves | IMP | Zfish | PMID:24741362 | |
| Biological Process | GO:0060047 | heart contraction | bves | IMP | ZFIN:ZDB-GENO-160323-2 | Zfish | PMID:26642364 |
| Biological Process | GO:0007507 | heart development | BVES | IMP | Human | PMID:26642364 | |
| Biological Process | GO:0007507 | heart development | bves | IMP | Zfish | PMID:26642364|ZFIN:ZDB-PUB-151208-1 | |
| Biological Process | GO:0001947 | heart looping | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:32843646 |
| Biological Process | GO:0002244 | hematopoietic progenitor cell differentiation | Bves | IGI | MGI:MGI:1343184 | Mouse | PMID:24029230 |
| Biological Process | GO:0003151 | outflow tract morphogenesis | bves | IMP | ZFIN:ZDB-MRPHLNO-130104-1 | Zfish | PMID:32843646 |
| Biological Process | GO:0040017 | positive regulation of locomotion | Bves | IDA | Mouse | MGI:MGI:4457243|PMID:18541910 | |
| Biological Process | GO:0001921 | positive regulation of receptor recycling | Bves | IDA | Mouse | MGI:MGI:4431297|PMID:20057356 | |
| Biological Process | GO:0008360 | regulation of cell shape | Bves | IDA | Mouse | MGI:MGI:4457243|PMID:18541910 | |
| Biological Process | GO:2001135 | regulation of endocytic recycling | Bves | IMP | Mouse | MGI:MGI:5613454|PMID:24048452 | |
| Biological Process | GO:0043087 | regulation of GTPase activity | Bves | IDA | Mouse | MGI:MGI:4457243|PMID:18541910 | |
| Biological Process | GO:0002027 | regulation of heart rate | Bves | IMP | MGI:MGI:2385915 | Mouse | PMID:22354168 |
| Biological Process | GO:0042391 | regulation of membrane potential | Bves | IGI | MGI:MGI:109366 | Mouse | PMID:22354168 |
| Biological Process | GO:0002931 | response to ischemia | Bves | IMP | Mouse | MGI:MGI:5550350|PMID:24066022 | |
| Biological Process | GO:0060041 | retina development in camera-type eye | bves | IMP | Zfish | PMID:24741362 | |
| Biological Process | GO:0010842 | retina layer formation | bves | IMP | Zfish | PMID:24741362 | |
| Biological Process | GO:0060931 | sinoatrial node cell development | Bves | IMP | MGI:MGI:2385915 | Mouse | PMID:22354168 |
| Biological Process | GO:0007519 | skeletal muscle tissue development | BVES | IMP | Human | PMID:26642364 | |
| Biological Process | GO:0007519 | skeletal muscle tissue development | bves | IMP | Zfish | PMID:26642364|ZFIN:ZDB-PUB-151208-1 | |
| Biological Process | GO:0007519 | skeletal muscle tissue development | bves | IMP | ZFIN:ZDB-MRPHLNO-160310-7,ZFIN:ZDB-MRPHLNO-160310-8 | Zfish | PMID:26642364 |
| Biological Process | GO:0014866 | skeletal myofibril assembly | bves | IMP | ZFIN:ZDB-GENO-160323-2 | Zfish | PMID:26642364 |
| Biological Process | GO:0014866 | skeletal myofibril assembly | bves | IMP | ZFIN:ZDB-MRPHLNO-160310-7,ZFIN:ZDB-MRPHLNO-160310-8 | Zfish | PMID:26642364 |
| Biological Process | GO:0034446 | substrate adhesion-dependent cell spreading | Bves | IDA | Mouse | MGI:MGI:4431297|PMID:20057356 | |
| Biological Process | GO:0048278 | vesicle docking | Bves | IMP | Mouse | MGI:MGI:5613454|PMID:24048452 | |
| Biological Process | GO:0016192 | vesicle-mediated transport | Bves | IDA | Mouse | MGI:MGI:4431297|PMID:20057356 |
| EXP Inferred from experiment |
| IDA Inferred from direct assay |
| IEP Inferred from expression pattern |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| HTP Inferred from High Throughput Experiment |
| HDA Inferred from High Throughput Direct Assay |
| HMP Inferred from High Throughput Mutant Phenotype |
| HGI Inferred from High Throughput Genetic Interaction |
| HEP Inferred from High Throughput Expression Pattern |
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||