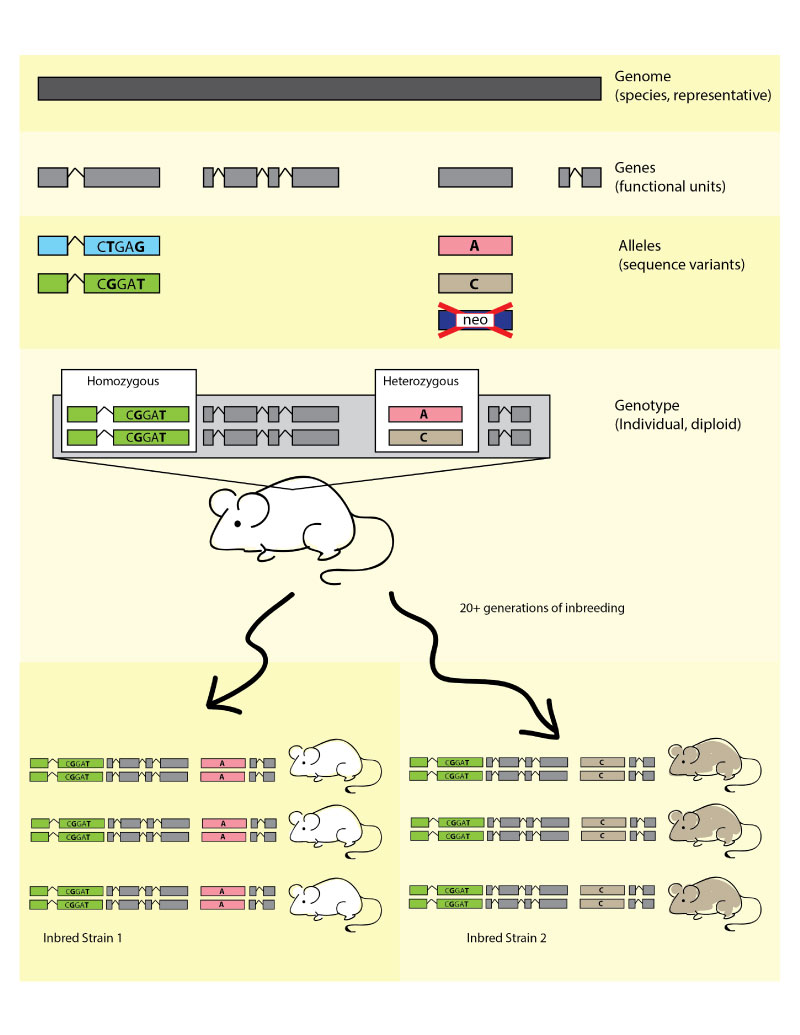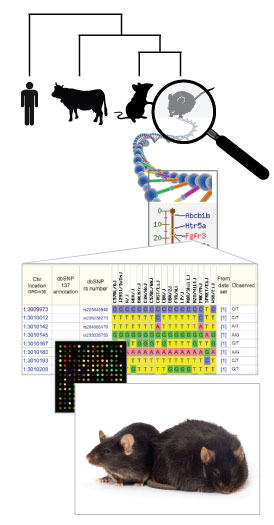
Figure 2. Infographic illustration of the hierarchy
between species level genomes, genes (functional units), the sequence variation
which defines alleles, leading to individual genotypes, which, when expressed
dictate phenotypes. See right for details. The bottom photo shows an obese C57BL/6J-
Lepob/Lepob mouse on the right, with a
heterozygous Lepob/Lep+ lean littermate
on the left.
|
Within a species, all members carry the same set of genes,
or functional units. The mouse reference genome contains an approximately
24,500 protein-coding genes and was fully sequenced in 2002
[2]. Structurally,
these are distributed across 19 pairs of autosomes, plus X and Y sex chromosomes.
For the sake of comparison, humans have 23 pairs of chromosomes (22 autosomes
plus XX or XY) which contain an estimated 20,687 protein-coding genes
[3]. Over 90% of the mouse and
human genomes can be aligned into regions of shared synteny, which is to say,
blocks where homologous genes are conserved in the same relative order,
indicative of shared common ancestry. At the gene level, a mouse homolog has
been identified and classified for
17,096 human genes.
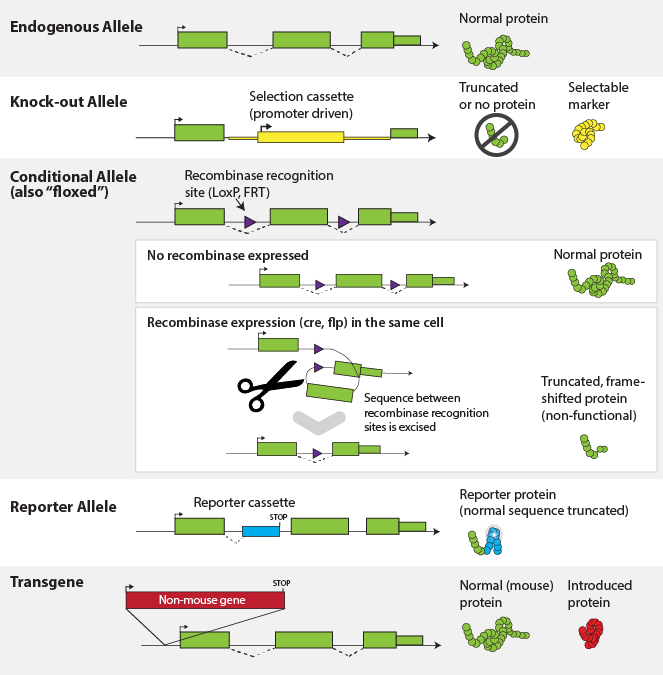
Variability between individuals within a species is due to variant gene
forms called alleles. Some changes are able to alter the expression or function
of genes resulting in observable phenotypic differences, such as coat color,
disease resistance/susceptibility, or metabolism but many other alleles result in
little to no detectable variation beyond the sequence level. Variant alleles may
include single nucleotide polymorphisms (SNPs), insertions and deletions (indels),
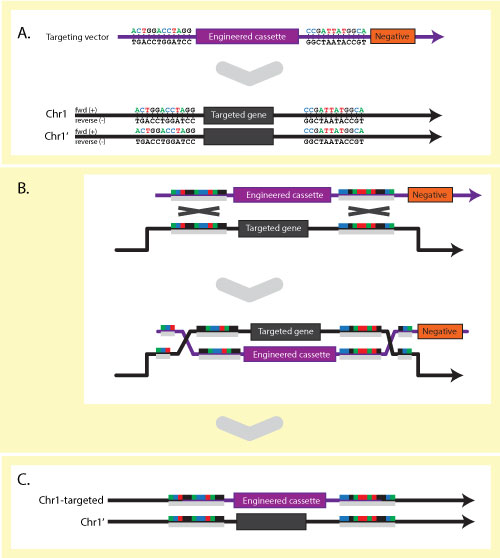
or copy number variation (CNVs). In the case of laboratory model organisms,
targeted alleles also exist where genetic engineering techniques have been used
to specifically alter, delete or introduce DNA sequences. A single gene may have
multiple alleles with different biological consequences, and every allele in MGI
is given a unique identifier, which is appended to the gene symbol as a superscript.
See the section below on Nomenclature for more.
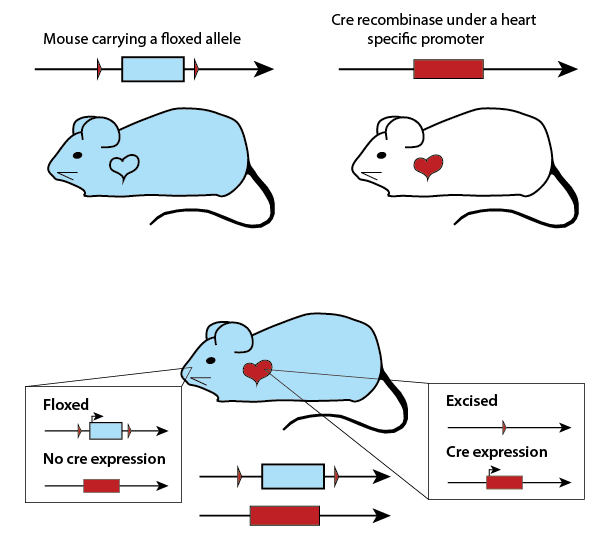
Barring the unusual case of chimeras, every nucleated cell in a mouse’s
body carries the same DNA code. However, different gene expression patterns allow
these cells to develop into a variety of tissues and organs, as well as respond
to stimuli. The
Gene Expression Database at MGI allows you to search for genes which are expressed
in different mouse anatomical structures, with a particular emphasis on embryonic
development.
The end result, whether a single measurement or the composite of all of
an individual's observable or measurable traits is called the phenotype. In MGI,
phenotypes are annotated to the complete genotype, which is comprised of both the
allele(s) of special interest as well as the background. Just as different
specific alleles may be expected to have different characteristics and biological
impacts (consider missense coding variants versus knockout alleles versus
conditional or reporter targeted alleles), other alleles in the strain background,
whether known or unknown, can have modifier effects on a trait, or set of traits.
For example, the
Lepob allele shows background sensitivity with homozygous
Lepob/Lepob
on a C57BL/6J background exhibiting severe obesity with a pre-diabetes-like
syndrome, while Lepob/Lepob
on a C57BLKS background become
severely diabetic and infertile in addition to obese, with a significantly shortened
life expectancy.
|
 Analysis Tools
Analysis Tools