|
Symbol Name ID |
Pcgf2
polycomb group ring finger 2 MGI:99161 |
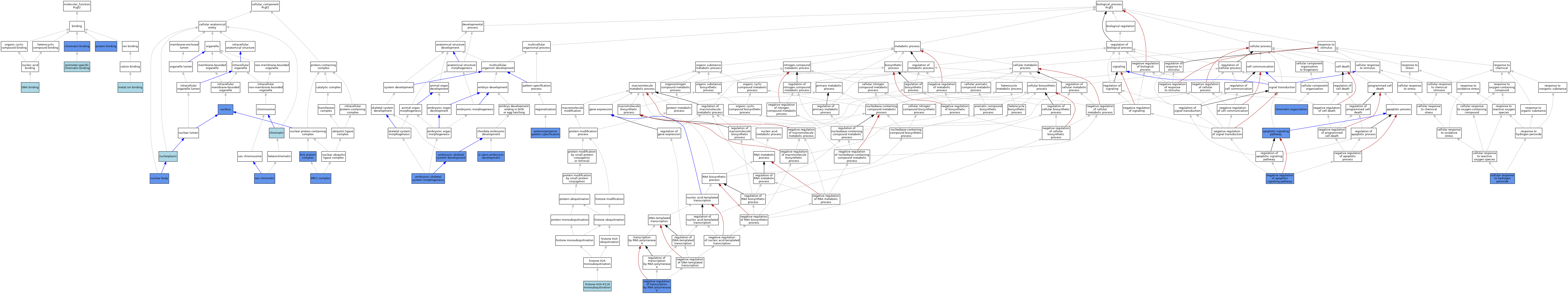
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:109525 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:96358 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:1990841 | promoter-specific chromatin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:186106 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:243420 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:100027 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:122905 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:104662 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:248225 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:186106 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:79851 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:77078 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:96358 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016604 | nuclear body | IDA | J:79851 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5619441 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:243420 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:77078 | |||||||||
| Cellular Component | GO:0031519 | PcG protein complex | IDA | J:243420 | |||||||||
| Cellular Component | GO:0031519 | PcG protein complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0035102 | PRC1 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0035102 | PRC1 complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0035102 | PRC1 complex | IDA | J:104662 | |||||||||
| Cellular Component | GO:0001739 | sex chromatin | IDA | J:94519 | |||||||||
| Biological Process | GO:0009952 | anterior/posterior pattern specification | IGI | J:96358 | |||||||||
| Biological Process | GO:0009952 | anterior/posterior pattern specification | IMP | J:33159 | |||||||||
| Biological Process | GO:0097190 | apoptotic signaling pathway | IGI | J:68216 | |||||||||
| Biological Process | GO:0070301 | cellular response to hydrogen peroxide | IDA | J:198334 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IGI | J:109525 | |||||||||
| Biological Process | GO:0048706 | embryonic skeletal system development | IGI | J:68216 | |||||||||
| Biological Process | GO:0048704 | embryonic skeletal system morphogenesis | IGI | J:68216 | |||||||||
| Biological Process | GO:0036353 | histone H2A-K119 monoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0036353 | histone H2A-K119 monoubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0001701 | in utero embryonic development | IGI | J:68216 | |||||||||
| Biological Process | GO:2001234 | negative regulation of apoptotic signaling pathway | IGI | J:68216 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:68216 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||