|
Symbol Name ID |
Pcsk1
proprotein convertase subtilisin/kexin type 1 MGI:97511 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
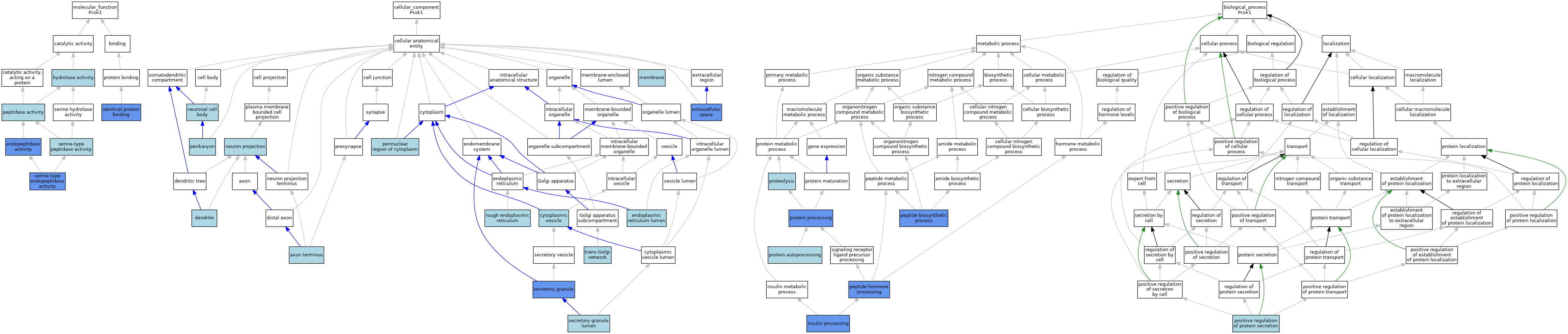
| Molecular Function | GO:0004175 | endopeptidase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0004175 | endopeptidase activity | IDA | J:139345 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | IPI | J:248247 | |||||||||
| Molecular Function | GO:0008233 | peptidase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IDA | J:131459 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | TAS | Reactome:R-MMU-216806 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | TAS | Reactome:R-MMU-508205 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | TAS | Reactome:R-MMU-508217 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | TAS | Reactome:R-MMU-508227 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0004252 | serine-type endopeptidase activity | IMP | J:78368 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0008236 | serine-type peptidase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043679 | axon terminus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0030425 | dendrite | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005788 | endoplasmic reticulum lumen | TAS | Reactome:R-MMU-181788 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:135486 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043005 | neuron projection | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043025 | neuronal cell body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043204 | perikaryon | ISO | J:155856 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005791 | rough endoplasmic reticulum | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030141 | secretory granule | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030141 | secretory granule | ISO | J:155856 | |||||||||
| Cellular Component | GO:0030141 | secretory granule | IDA | J:78368 | |||||||||
| Cellular Component | GO:0034774 | secretory granule lumen | TAS | Reactome:R-MMU-216806 | |||||||||
| Cellular Component | GO:0034774 | secretory granule lumen | TAS | Reactome:R-MMU-508205 | |||||||||
| Cellular Component | GO:0034774 | secretory granule lumen | TAS | Reactome:R-MMU-508217 | |||||||||
| Cellular Component | GO:0034774 | secretory granule lumen | TAS | Reactome:R-MMU-508227 | |||||||||
| Cellular Component | GO:0005802 | trans-Golgi network | ISO | J:155856 | |||||||||
| Biological Process | GO:0030070 | insulin processing | IMP | J:78368 | |||||||||
| Biological Process | GO:0043043 | peptide biosynthetic process | IDA | J:131459 | |||||||||
| Biological Process | GO:0016486 | peptide hormone processing | IDA | J:139345 | |||||||||
| Biological Process | GO:0016486 | peptide hormone processing | ISO | J:155856 | |||||||||
| Biological Process | GO:0016486 | peptide hormone processing | IDA | J:131459 | |||||||||
| Biological Process | GO:0050714 | positive regulation of protein secretion | ISO | J:155856 | |||||||||
| Biological Process | GO:0016540 | protein autoprocessing | ISO | J:155856 | |||||||||
| Biological Process | GO:0016485 | protein processing | IDA | J:139345 | |||||||||
| Biological Process | GO:0016485 | protein processing | ISO | J:155856 | |||||||||
| Biological Process | GO:0016485 | protein processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:60000 | |||||||||
| Biological Process | GO:0006508 | proteolysis | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||