|
Symbol Name ID |
Atp5f1a
ATP synthase F1 subunit alpha MGI:88115 |
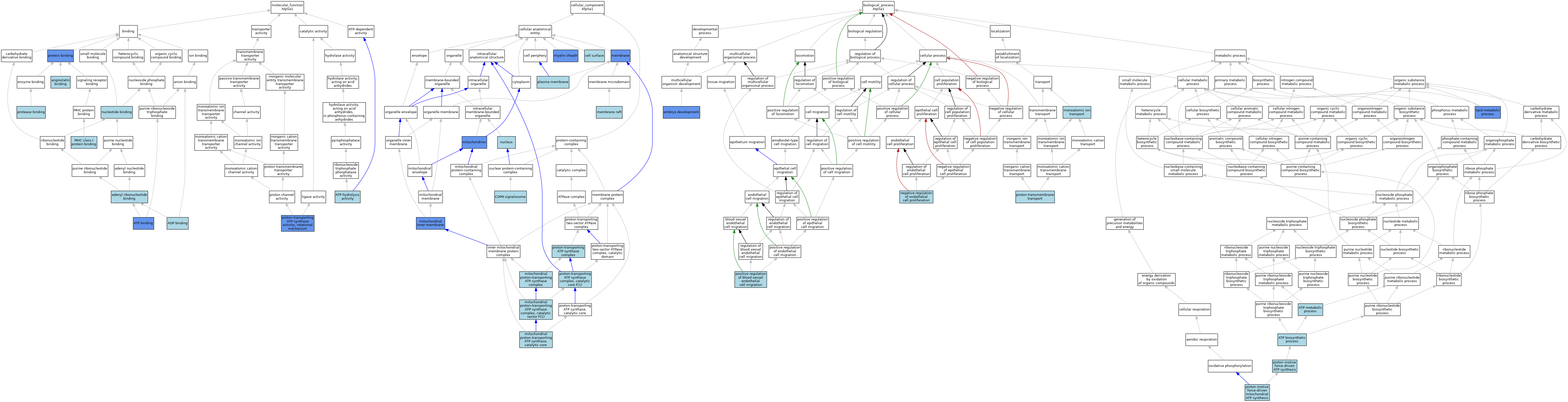
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0032559 | adenyl ribonucleotide binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0043531 | ADP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0043531 | ADP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0043532 | angiostatin binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IMP | J:4125 | |||||||||
| Molecular Function | GO:0016887 | ATP hydrolysis activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042288 | MHC class I protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0002020 | protease binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:235261 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:313842 | |||||||||
| Molecular Function | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism | IMP | J:123729 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:155856 | |||||||||
| Cellular Component | GO:0008180 | COP9 signalosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | IMP | J:156519 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045121 | membrane raft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005743 | mitochondrial inner membrane | HDA | J:100953 | |||||||||
| Cellular Component | GO:0005753 | mitochondrial proton-transporting ATP synthase complex | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005753 | mitochondrial proton-transporting ATP synthase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic sector F(1) | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005754 | mitochondrial proton-transporting ATP synthase, catalytic core | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:153657 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:223336 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | IDA | J:233904 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:86816 | |||||||||
| Cellular Component | GO:0043209 | myelin sheath | HDA | J:145263 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045259 | proton-transporting ATP synthase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1) | ISO | J:155856 | |||||||||
| Cellular Component | GO:0045261 | proton-transporting ATP synthase complex, catalytic core F(1) | IBA | J:265628 | |||||||||
| Biological Process | GO:0006754 | ATP biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0046034 | ATP metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0009790 | embryo development | IMP | J:122207 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IMP | J:123729 | |||||||||
| Biological Process | GO:0006811 | monoatomic ion transport | IEA | J:60000 | |||||||||
| Biological Process | GO:0001937 | negative regulation of endothelial cell proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043536 | positive regulation of blood vessel endothelial cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0015986 | proton motive force-driven ATP synthesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0042776 | proton motive force-driven mitochondrial ATP synthesis | ISO | J:164563 | |||||||||
| Biological Process | GO:1902600 | proton transmembrane transport | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||