|
Symbol Name ID |
Areg
amphiregulin MGI:88068 |
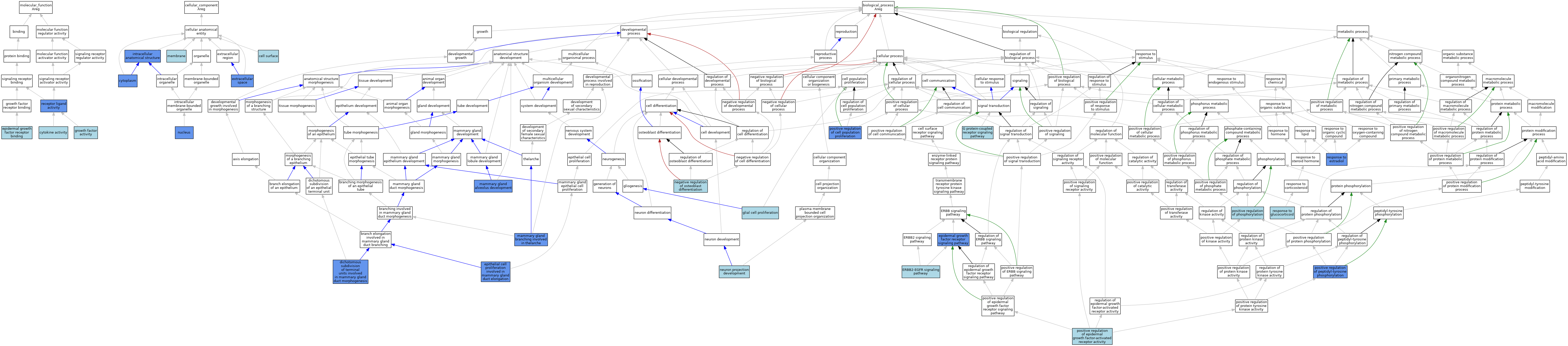
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005125 | cytokine activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005154 | epidermal growth factor receptor binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008083 | growth factor activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IGI | J:226559 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IGI | J:193394 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IGI | J:277693 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | IGI | J:193417 | |||||||||
| Molecular Function | GO:0048018 | receptor ligand activity | ISO | J:34905 | |||||||||
| Cellular Component | GO:0009986 | cell surface | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:27180 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:193417 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IGI | J:277693 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:34905 | |||||||||
| Cellular Component | GO:0005622 | intracellular anatomical structure | IDA | J:27180 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:27180 | |||||||||
| Biological Process | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis | IMP | J:120116 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | ISO | J:155856 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IGI | J:193394 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IGI | J:193417 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IGI | J:277693 | |||||||||
| Biological Process | GO:0007173 | epidermal growth factor receptor signaling pathway | IGI | J:226559 | |||||||||
| Biological Process | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation | IMP | J:120116 | |||||||||
| Biological Process | GO:0038134 | ERBB2-EGFR signaling pathway | ISO | J:34905 | |||||||||
| Biological Process | GO:0007186 | G protein-coupled receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0014009 | glial cell proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0060749 | mammary gland alveolus development | IGI | J:54980 | |||||||||
| Biological Process | GO:0060749 | mammary gland alveolus development | IGI | J:54980 | |||||||||
| Biological Process | GO:0060744 | mammary gland branching involved in thelarche | IMP | J:54980 | |||||||||
| Biological Process | GO:0060744 | mammary gland branching involved in thelarche | IMP | J:120116 | |||||||||
| Biological Process | GO:0045668 | negative regulation of osteoblast differentiation | ISO | J:155856 | |||||||||
| Biological Process | GO:0031175 | neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:155856 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IBA | J:265628 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | IDA | J:34905 | |||||||||
| Biological Process | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity | IBA | J:265628 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | IDA | J:34905 | |||||||||
| Biological Process | GO:0042327 | positive regulation of phosphorylation | ISO | J:155856 | |||||||||
| Biological Process | GO:0032355 | response to estradiol | IDA | J:120116 | |||||||||
| Biological Process | GO:0051384 | response to glucocorticoid | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||