|
Symbol Name ID |
Rbm15
RNA binding motif protein 15 MGI:2443205 |
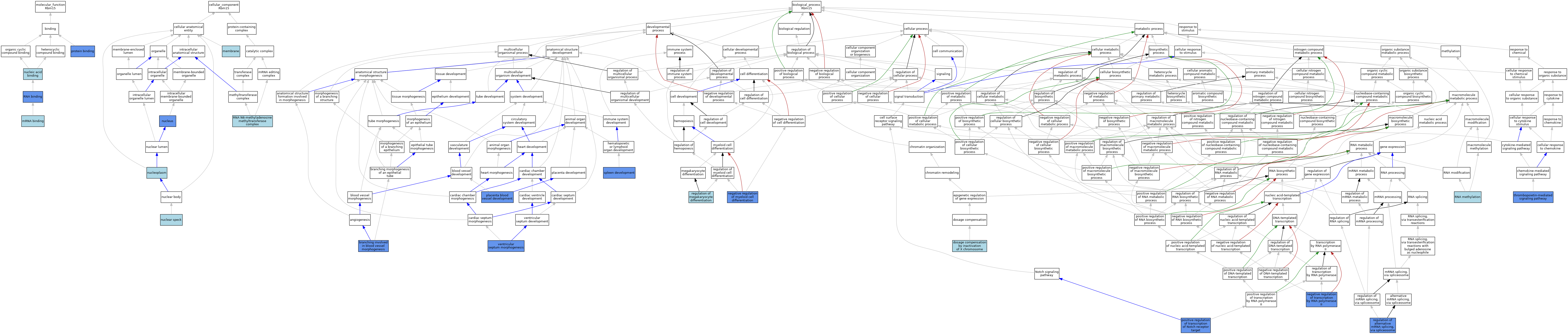
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003729 | mRNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003729 | mRNA binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0003676 | nucleic acid binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:121406 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IDA | J:261836 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0016607 | nuclear speck | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:121406 | |||||||||
| Cellular Component | GO:0036396 | RNA N6-methyladenosine methyltransferase complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0001569 | branching involved in blood vessel morphogenesis | IMP | J:145030 | |||||||||
| Biological Process | GO:0009048 | dosage compensation by inactivation of X chromosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0045638 | negative regulation of myeloid cell differentiation | IMP | J:121406 | |||||||||
| Biological Process | GO:0045638 | negative regulation of myeloid cell differentiation | IDA | J:121406 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IDA | J:121406 | |||||||||
| Biological Process | GO:0060674 | placenta blood vessel development | IMP | J:145030 | |||||||||
| Biological Process | GO:0007221 | positive regulation of transcription of Notch receptor target | IDA | J:121406 | |||||||||
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | IDA | J:261836 | |||||||||
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | ISO | J:164563 | |||||||||
| Biological Process | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome | IBA | J:265628 | |||||||||
| Biological Process | GO:0045652 | regulation of megakaryocyte differentiation | ISO | J:164563 | |||||||||
| Biological Process | GO:0001510 | RNA methylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0048536 | spleen development | IMP | J:145030 | |||||||||
| Biological Process | GO:0038163 | thrombopoietin-mediated signaling pathway | IDA | J:261836 | |||||||||
| Biological Process | GO:0060412 | ventricular septum morphogenesis | IMP | J:145030 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||