|
Symbol Name ID |
Hif1an
hypoxia-inducible factor 1, alpha subunit inhibitor MGI:2442345 |
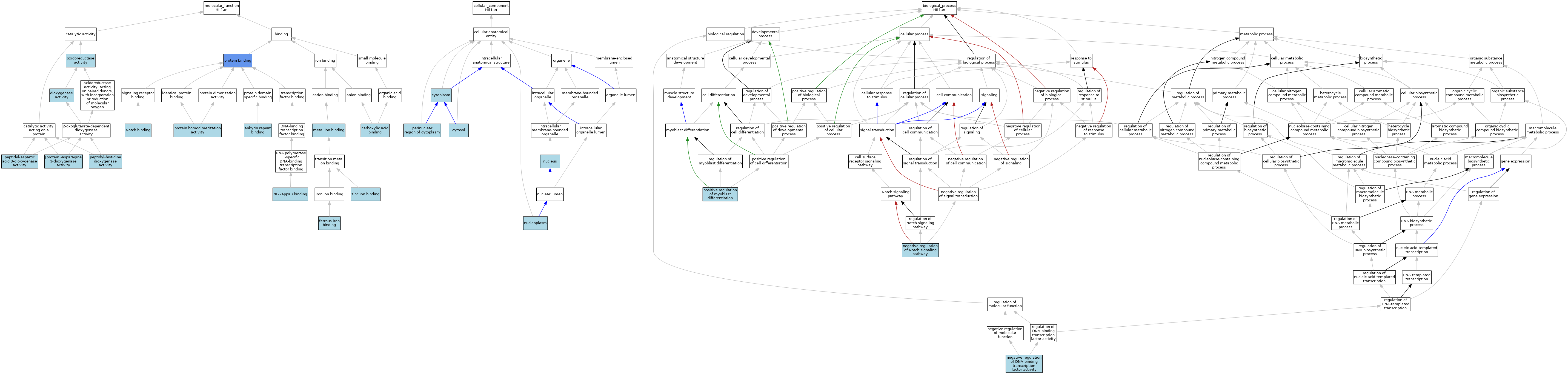
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0036140 | [protein]-asparagine 3-dioxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0036140 | [protein]-asparagine 3-dioxygenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0071532 | ankyrin repeat binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0071532 | ankyrin repeat binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031406 | carboxylic acid binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051213 | dioxygenase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008198 | ferrous iron binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0051059 | NF-kappaB binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005112 | Notch binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016491 | oxidoreductase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0062101 | peptidyl-aspartic acid 3-dioxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0036139 | peptidyl-histidine dioxygenase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0036139 | peptidyl-histidine dioxygenase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:219557 | |||||||||
| Molecular Function | GO:0042803 | protein homodimerization activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0043433 | negative regulation of DNA-binding transcription factor activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0045746 | negative regulation of Notch signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0045746 | negative regulation of Notch signaling pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0045663 | positive regulation of myoblast differentiation | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||