|
Symbol Name ID |
Nup153
nucleoporin 153 MGI:2385621 |
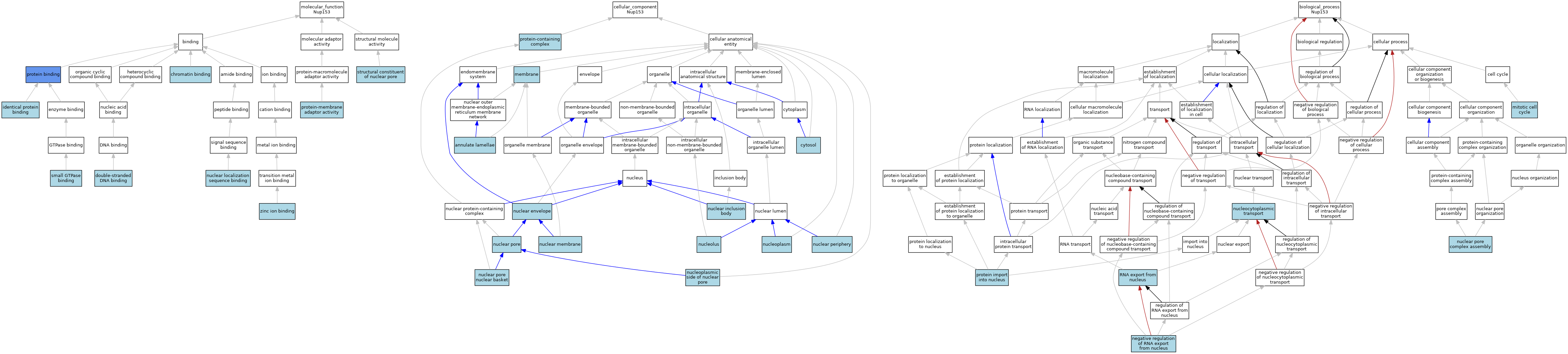
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0003690 | double-stranded DNA binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008139 | nuclear localization sequence binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008139 | nuclear localization sequence binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:63241 | |||||||||
| Molecular Function | GO:0043495 | protein-membrane adaptor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031267 | small GTPase binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0017056 | structural constituent of nuclear pore | ISO | J:164563 | |||||||||
| Molecular Function | GO:0017056 | structural constituent of nuclear pore | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005642 | annulate lamellae | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005635 | nuclear envelope | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042405 | nuclear inclusion body | ISO | J:155856 | |||||||||
| Cellular Component | GO:0042405 | nuclear inclusion body | ISO | J:164563 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0031965 | nuclear membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0034399 | nuclear periphery | ISO | J:155856 | |||||||||
| Cellular Component | GO:0034399 | nuclear periphery | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | NAS | J:320054 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005643 | nuclear pore | IBA | J:265628 | |||||||||
| Cellular Component | GO:0044615 | nuclear pore nuclear basket | ISO | J:164563 | |||||||||
| Cellular Component | GO:0044615 | nuclear pore nuclear basket | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990875 | nucleoplasmic side of nuclear pore | ISO | J:155856 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | ISO | J:155856 | |||||||||
| Biological Process | GO:0000278 | mitotic cell cycle | ISO | J:155856 | |||||||||
| Biological Process | GO:0046832 | negative regulation of RNA export from nucleus | ISO | J:164563 | |||||||||
| Biological Process | GO:0051292 | nuclear pore complex assembly | ISO | J:164563 | |||||||||
| Biological Process | GO:0006913 | nucleocytoplasmic transport | NAS | J:320054 | |||||||||
| Biological Process | GO:0006606 | protein import into nucleus | IBA | J:265628 | |||||||||
| Biological Process | GO:0006405 | RNA export from nucleus | IBA | J:265628 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||