|
Symbol Name ID |
Pla2g15
phospholipase A2, group XV MGI:2178076 |
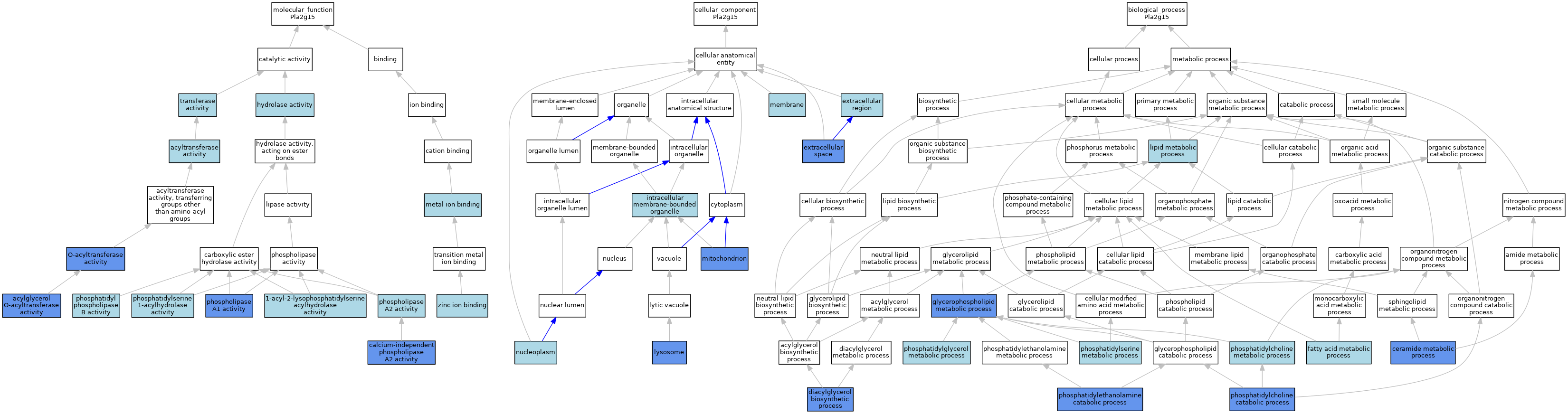
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0016411 | acylglycerol O-acyltransferase activity | IDA | J:284981 | |||||||||
| Molecular Function | GO:0016746 | acyltransferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | IDA | J:284981 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | IDA | J:116483 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0047499 | calcium-independent phospholipase A2 activity | IDA | J:75456 | |||||||||
| Molecular Function | GO:0016787 | hydrolase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008374 | O-acyltransferase activity | IDA | J:162943 | |||||||||
| Molecular Function | GO:0008374 | O-acyltransferase activity | IDA | J:116483 | |||||||||
| Molecular Function | GO:0008374 | O-acyltransferase activity | ISO | J:155856 | |||||||||
| Molecular Function | GO:0008374 | O-acyltransferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008374 | O-acyltransferase activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0102545 | phosphatidyl phospholipase B activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0052739 | phosphatidylserine 1-acylhydrolase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0008970 | phospholipase A1 activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0008970 | phospholipase A1 activity | IDA | J:116483 | |||||||||
| Molecular Function | GO:0004623 | phospholipase A2 activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IDA | J:162943 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | HDA | J:221550 | |||||||||
| Cellular Component | GO:0005615 | extracellular space | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005764 | lysosome | IDA | J:261411 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005739 | mitochondrion | HDA | J:151002 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | IDA | J:116483 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | IDA | J:162943 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0006672 | ceramide metabolic process | IDA | J:75456 | |||||||||
| Biological Process | GO:0006651 | diacylglycerol biosynthetic process | IDA | J:284981 | |||||||||
| Biological Process | GO:0006631 | fatty acid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0006650 | glycerophospholipid metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006650 | glycerophospholipid metabolic process | IDA | J:162943 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:72247 | |||||||||
| Biological Process | GO:0006629 | lipid metabolic process | IEA | J:60000 | |||||||||
| Biological Process | GO:0034638 | phosphatidylcholine catabolic process | IMP | J:111561 | |||||||||
| Biological Process | GO:0034638 | phosphatidylcholine catabolic process | IDA | J:284981 | |||||||||
| Biological Process | GO:0034638 | phosphatidylcholine catabolic process | IDA | J:116483 | |||||||||
| Biological Process | GO:0046470 | phosphatidylcholine metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0046470 | phosphatidylcholine metabolic process | ISO | J:155856 | |||||||||
| Biological Process | GO:0046470 | phosphatidylcholine metabolic process | IBA | J:265628 | |||||||||
| Biological Process | GO:0046338 | phosphatidylethanolamine catabolic process | IMP | J:111561 | |||||||||
| Biological Process | GO:0046338 | phosphatidylethanolamine catabolic process | IDA | J:116483 | |||||||||
| Biological Process | GO:0046471 | phosphatidylglycerol metabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006658 | phosphatidylserine metabolic process | ISO | J:164563 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||