|
Symbol Name ID |
Cspg4
chondroitin sulfate proteoglycan 4 MGI:2153093 |
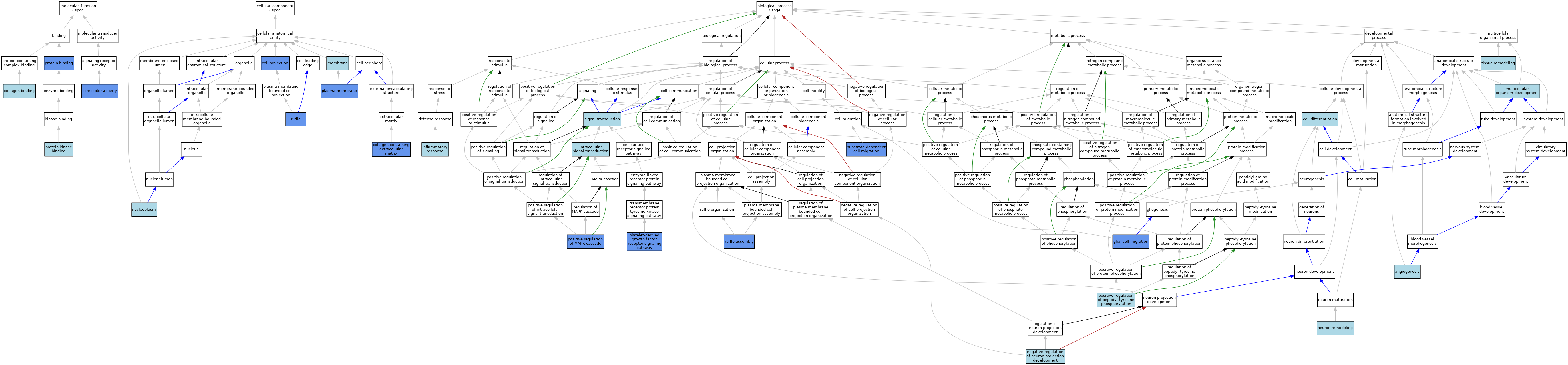
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005518 | collagen binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0015026 | coreceptor activity | IDA | J:227201 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:81682 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:227201 | |||||||||
| Molecular Function | GO:0019901 | protein kinase binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IDA | J:227201 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IDA | J:196061 | |||||||||
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265604 | |||||||||
| Cellular Component | GO:0062023 | collagen-containing extracellular matrix | HDA | J:265604 | |||||||||
| Cellular Component | GO:0016020 | membrane | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:71947 | |||||||||
| Cellular Component | GO:0001726 | ruffle | IDA | J:227201 | |||||||||
| Biological Process | GO:0001525 | angiogenesis | IEA | J:60000 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0008347 | glial cell migration | IDA | J:76139 | |||||||||
| Biological Process | GO:0006954 | inflammatory response | ISO | J:155856 | |||||||||
| Biological Process | GO:0035556 | intracellular signal transduction | ISO | J:164563 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0010977 | negative regulation of neuron projection development | ISO | J:155856 | |||||||||
| Biological Process | GO:0016322 | neuron remodeling | ISO | J:155856 | |||||||||
| Biological Process | GO:0048008 | platelet-derived growth factor receptor signaling pathway | IDA | J:227201 | |||||||||
| Biological Process | GO:0043410 | positive regulation of MAPK cascade | IMP | J:54319 | |||||||||
| Biological Process | GO:0050731 | positive regulation of peptidyl-tyrosine phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0097178 | ruffle assembly | IDA | J:227201 | |||||||||
| Biological Process | GO:0007165 | signal transduction | IEA | J:60000 | |||||||||
| Biological Process | GO:0006929 | substrate-dependent cell migration | IGI | J:227201 | |||||||||
| Biological Process | GO:0048771 | tissue remodeling | IEA | J:60000 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||