|
Symbol Name ID |
Mael
maelstrom spermatogenic transposon silencer MGI:2138453 |
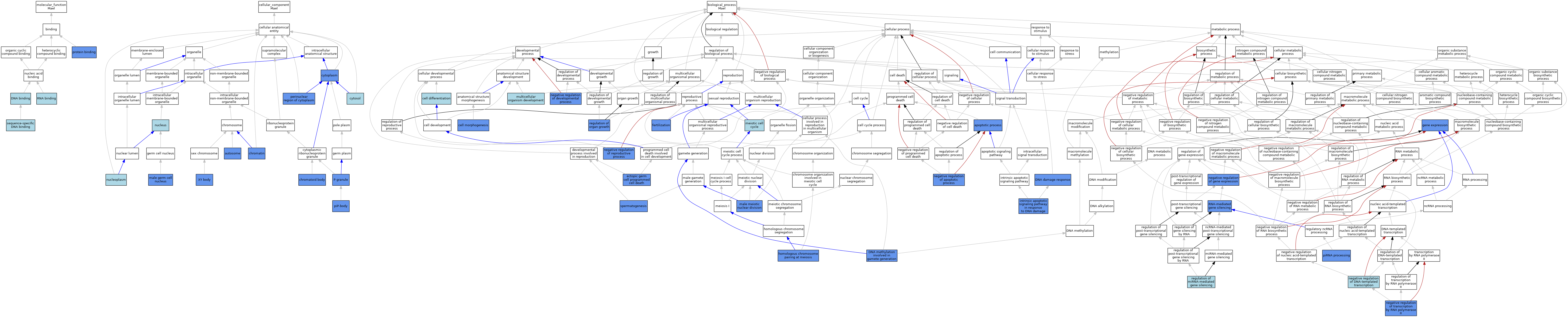
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:243444 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:112036 | |||||||||
| Molecular Function | GO:0003723 | RNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0043565 | sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030849 | autosome | IDA | J:112036 | |||||||||
| Cellular Component | GO:0000785 | chromatin | IDA | J:112036 | |||||||||
| Cellular Component | GO:0033391 | chromatoid body | IDA | J:112036 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:140402 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:112036 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IDA | J:112036 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5606210 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5606252 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5606256 | |||||||||
| Cellular Component | GO:0005829 | cytosol | TAS | Reactome:R-MMU-5629230 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:112036 | |||||||||
| Cellular Component | GO:0001673 | male germ cell nucleus | IDA | J:140402 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | TAS | Reactome:R-MMU-5606256 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043186 | P granule | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043186 | P granule | IDA | J:112036 | |||||||||
| Cellular Component | GO:0043186 | P granule | IDA | J:140402 | |||||||||
| Cellular Component | GO:0048471 | perinuclear region of cytoplasm | IDA | J:140402 | |||||||||
| Cellular Component | GO:0071547 | piP-body | IDA | J:155865 | |||||||||
| Cellular Component | GO:0001741 | XY body | IDA | J:112036 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | IMP | J:140402 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0000902 | cell morphogenesis | IMP | J:140402 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IMP | J:140402 | |||||||||
| Biological Process | GO:0043046 | DNA methylation involved in gamete generation | IMP | J:155865 | |||||||||
| Biological Process | GO:0035234 | ectopic germ cell programmed cell death | IMP | J:140402 | |||||||||
| Biological Process | GO:0009566 | fertilization | IMP | J:140402 | |||||||||
| Biological Process | GO:0010467 | gene expression | IMP | J:140402 | |||||||||
| Biological Process | GO:0007129 | homologous chromosome pairing at meiosis | IMP | J:140402 | |||||||||
| Biological Process | GO:0007129 | homologous chromosome pairing at meiosis | IMP | J:140402 | |||||||||
| Biological Process | GO:0008630 | intrinsic apoptotic signaling pathway in response to DNA damage | IMP | J:140402 | |||||||||
| Biological Process | GO:0007140 | male meiotic nuclear division | IBA | J:265628 | |||||||||
| Biological Process | GO:0007140 | male meiotic nuclear division | IMP | J:140402 | |||||||||
| Biological Process | GO:0051321 | meiotic cell cycle | IEA | J:60000 | |||||||||
| Biological Process | GO:0007275 | multicellular organism development | IEA | J:60000 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IMP | J:140402 | |||||||||
| Biological Process | GO:0051093 | negative regulation of developmental process | IMP | J:140402 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IBA | J:265628 | |||||||||
| Biological Process | GO:0010629 | negative regulation of gene expression | IMP | J:140402 | |||||||||
| Biological Process | GO:2000242 | negative regulation of reproductive process | IMP | J:140402 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IMP | J:140402 | |||||||||
| Biological Process | GO:0034587 | piRNA processing | IMP | J:155865 | |||||||||
| Biological Process | GO:0034587 | piRNA processing | IMP | J:140402 | |||||||||
| Biological Process | GO:0034587 | piRNA processing | IBA | J:265628 | |||||||||
| Biological Process | GO:0060964 | regulation of miRNA-mediated gene silencing | IEA | J:72247 | |||||||||
| Biological Process | GO:0046620 | regulation of organ growth | IMP | J:140402 | |||||||||
| Biological Process | GO:0031047 | RNA-mediated gene silencing | IMP | J:140402 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IBA | J:265628 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:140402 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||