|
Symbol Name ID |
Marchf7
membrane associated ring-CH-type finger 7 MGI:1931053 |
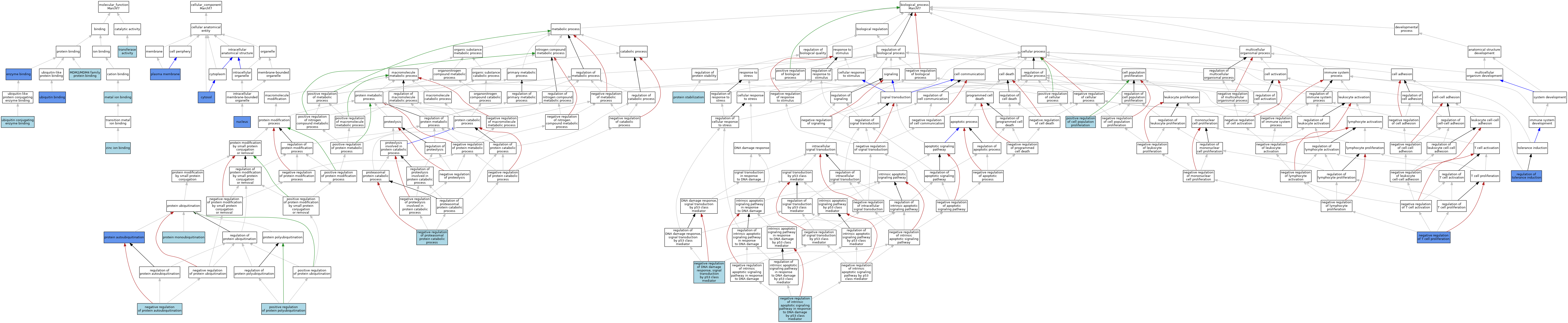
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0019899 | enzyme binding | IPI | J:262357 | |||||||||
| Molecular Function | GO:0097371 | MDM2/MDM4 family protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0043130 | ubiquitin binding | IMP | J:262357 | |||||||||
| Molecular Function | GO:0043130 | ubiquitin binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031624 | ubiquitin conjugating enzyme binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031624 | ubiquitin conjugating enzyme binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | IEA | J:72247 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:262357 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:262357 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005886 | plasma membrane | IDA | J:262357 | |||||||||
| Biological Process | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator | ISO | J:164563 | |||||||||
| Biological Process | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | ISO | J:164563 | |||||||||
| Biological Process | GO:1901799 | negative regulation of proteasomal protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:1905524 | negative regulation of protein autoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0042130 | negative regulation of T cell proliferation | IMP | J:96167 | |||||||||
| Biological Process | GO:0008284 | positive regulation of cell population proliferation | ISO | J:164563 | |||||||||
| Biological Process | GO:1902916 | positive regulation of protein polyubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | IMP | J:262357 | |||||||||
| Biological Process | GO:0051865 | protein autoubiquitination | IBA | J:265628 | |||||||||
| Biological Process | GO:0006513 | protein monoubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | ISO | J:164563 | |||||||||
| Biological Process | GO:0002643 | regulation of tolerance induction | IMP | J:96167 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 05/07/2024 MGI 6.23 |

|
|
|
||