|
Symbol Name ID |
Trio
triple functional domain (PTPRF interacting) MGI:1927230 |
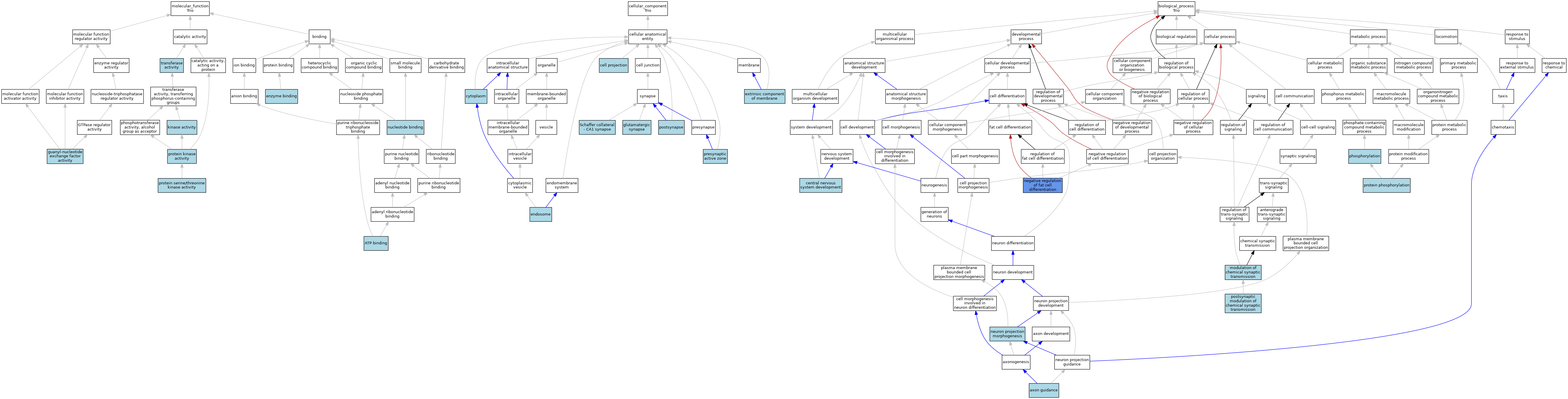
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005524 | ATP binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005524 | ATP binding | IEA | J:72247 | |||||||||
| Molecular Function | GO:0019899 | enzyme binding | ISO | J:155856 | |||||||||
| Molecular Function | GO:0005085 | guanyl-nucleotide exchange factor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0016301 | kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000166 | nucleotide binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004672 | protein kinase activity | IEA | J:72247 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0004674 | protein serine/threonine kinase activity | IEA | J:72245 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Cellular Component | GO:0042995 | cell projection | IEA | J:60000 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005768 | endosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0019898 | extrinsic component of membrane | IBA | J:265628 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0098978 | glutamatergic synapse | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098794 | postsynapse | ISO | J:155856 | |||||||||
| Cellular Component | GO:0048786 | presynaptic active zone | ISO | J:164563 | |||||||||
| Cellular Component | GO:0098685 | Schaffer collateral - CA1 synapse | ISO | J:155856 | |||||||||
| Biological Process | GO:0007411 | axon guidance | IBA | J:265628 | |||||||||
| Biological Process | GO:0007417 | central nervous system development | IEA | J:72247 | |||||||||
| Biological Process | GO:0050804 | modulation of chemical synaptic transmission | ISO | J:155856 | |||||||||
| Biological Process | GO:0045599 | negative regulation of fat cell differentiation | IMP | J:243390 | |||||||||
| Biological Process | GO:0048812 | neuron projection morphogenesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0016310 | phosphorylation | IEA | J:60000 | |||||||||
| Biological Process | GO:0099170 | postsynaptic modulation of chemical synaptic transmission | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | IEA | J:72247 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||