|
Symbol Name ID |
Uvrag
UV radiation resistance associated gene MGI:1925860 |
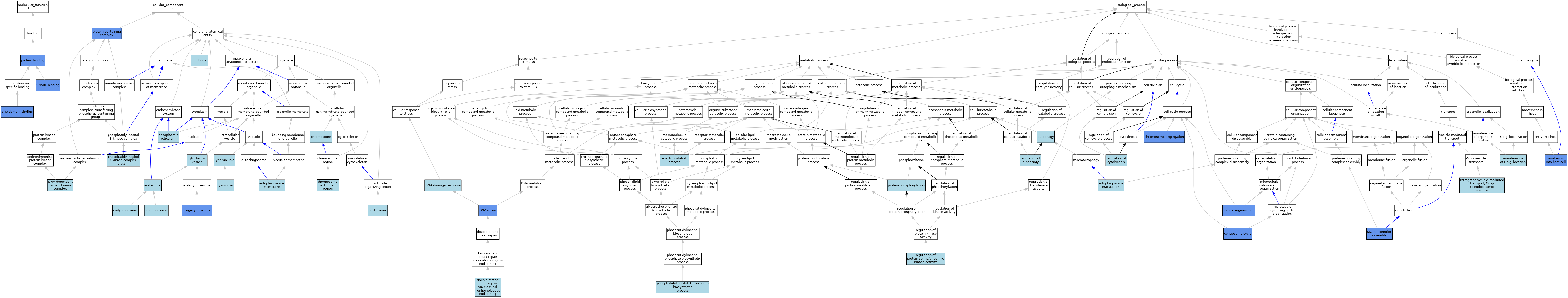
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005515 | protein binding | IPI | J:206800 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:240718 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:206800 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:206800 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:129990 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:185616 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:129990 | |||||||||
| Molecular Function | GO:0017124 | SH3 domain binding | IPI | J:129990 | |||||||||
| Molecular Function | GO:0000149 | SNARE binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0000149 | SNARE binding | IDA | J:206800 | |||||||||
| Cellular Component | GO:0000421 | autophagosome membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005813 | centrosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005694 | chromosome | IEA | J:60000 | |||||||||
| Cellular Component | GO:0000775 | chromosome, centromeric region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0031410 | cytoplasmic vesicle | IEA | J:60000 | |||||||||
| Cellular Component | GO:0070418 | DNA-dependent protein kinase complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005769 | early endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005783 | endoplasmic reticulum | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005768 | endosome | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005770 | late endosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005764 | lysosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000323 | lytic vacuole | IBA | J:265628 | |||||||||
| Cellular Component | GO:0030496 | midbody | ISO | J:164563 | |||||||||
| Cellular Component | GO:0045335 | phagocytic vesicle | IPI | J:185616 | |||||||||
| Cellular Component | GO:0035032 | phosphatidylinositol 3-kinase complex, class III | ISO | J:164563 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | IDA | J:129990 | |||||||||
| Biological Process | GO:0097352 | autophagosome maturation | ISO | J:164563 | |||||||||
| Biological Process | GO:0006914 | autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0007098 | centrosome cycle | ISO | J:164563 | |||||||||
| Biological Process | GO:0007098 | centrosome cycle | IMP | J:222146 | |||||||||
| Biological Process | GO:0007059 | chromosome segregation | IMP | J:222146 | |||||||||
| Biological Process | GO:0006974 | DNA damage response | IEA | J:60000 | |||||||||
| Biological Process | GO:0006281 | DNA repair | IMP | J:222146 | |||||||||
| Biological Process | GO:0006281 | DNA repair | ISO | J:164563 | |||||||||
| Biological Process | GO:0097680 | double-strand break repair via classical nonhomologous end joining | ISO | J:164563 | |||||||||
| Biological Process | GO:0051684 | maintenance of Golgi location | ISO | J:164563 | |||||||||
| Biological Process | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006468 | protein phosphorylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0032801 | receptor catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0010506 | regulation of autophagy | ISO | J:164563 | |||||||||
| Biological Process | GO:0032465 | regulation of cytokinesis | ISO | J:164563 | |||||||||
| Biological Process | GO:0071900 | regulation of protein serine/threonine kinase activity | ISO | J:164563 | |||||||||
| Biological Process | GO:0006890 | retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | ISO | J:164563 | |||||||||
| Biological Process | GO:0035493 | SNARE complex assembly | IBA | J:265628 | |||||||||
| Biological Process | GO:0035493 | SNARE complex assembly | IMP | J:206800 | |||||||||
| Biological Process | GO:0007051 | spindle organization | IMP | J:222146 | |||||||||
| Biological Process | GO:0046718 | viral entry into host cell | IMP | J:206800 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/30/2024 MGI 6.23 |

|
|
|
||