|
Symbol Name ID |
Bag6
BCL2-associated athanogene 6 MGI:1919439 |
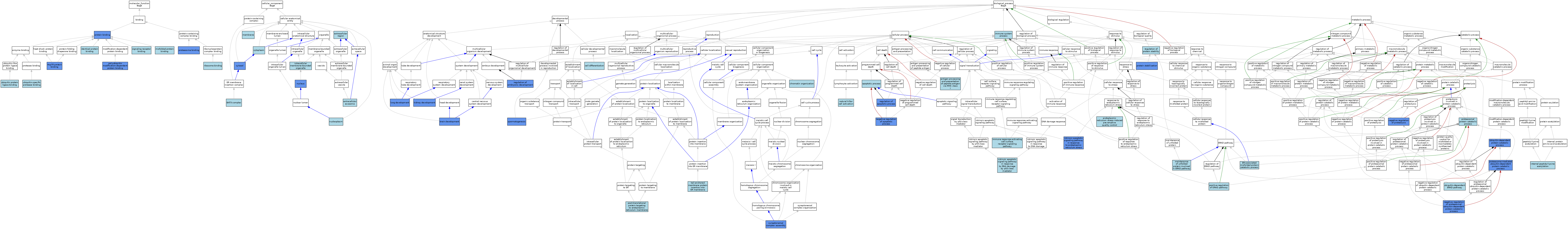
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0030544 | Hsp70 protein binding | IPI | J:139505 | |||||||||
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051787 | misfolded protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0051787 | misfolded protein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | IDA | J:168161 | |||||||||
| Molecular Function | GO:0031593 | polyubiquitin modification-dependent protein binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0070628 | proteasome binding | IDA | J:168161 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:131590 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:114108 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:182961 | |||||||||
| Molecular Function | GO:0043022 | ribosome binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0005102 | signaling receptor binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0031625 | ubiquitin protein ligase binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:1990381 | ubiquitin-specific protease binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0071818 | BAT3 complex | ISO | J:164563 | |||||||||
| Cellular Component | GO:0071818 | BAT3 complex | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISS | J:55895 | |||||||||
| Cellular Component | GO:0005829 | cytosol | NAS | J:320131 | |||||||||
| Cellular Component | GO:0005829 | cytosol | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005829 | cytosol | IDA | J:131590 | |||||||||
| Cellular Component | GO:0070062 | extracellular exosome | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005576 | extracellular region | IEA | J:60000 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0016020 | membrane | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IDA | J:182961 | |||||||||
| Biological Process | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I | TAS | J:168161 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006915 | apoptotic process | RCA | J:84168 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:119857 | |||||||||
| Biological Process | GO:0030154 | cell differentiation | IEA | J:60000 | |||||||||
| Biological Process | GO:0006325 | chromatin organization | IEA | J:60000 | |||||||||
| Biological Process | GO:0061857 | endoplasmic reticulum stress-induced pre-emptive quality control | ISO | J:164563 | |||||||||
| Biological Process | GO:0071712 | ER-associated misfolded protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0002429 | immune response-activating cell surface receptor signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0018393 | internal peptidyl-lysine acetylation | ISO | J:164563 | |||||||||
| Biological Process | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator | ISO | J:164563 | |||||||||
| Biological Process | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress | IMP | J:131590 | |||||||||
| Biological Process | GO:0001822 | kidney development | IMP | J:119857 | |||||||||
| Biological Process | GO:0030324 | lung development | IMP | J:119857 | |||||||||
| Biological Process | GO:1904378 | maintenance of unfolded protein involved in ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0030101 | natural killer cell activation | ISO | J:164563 | |||||||||
| Biological Process | GO:0043066 | negative regulation of apoptotic process | IGI | J:182961 | |||||||||
| Biological Process | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | IMP | J:139505 | |||||||||
| Biological Process | GO:0045861 | negative regulation of proteolysis | IMP | J:131590 | |||||||||
| Biological Process | GO:1904294 | positive regulation of ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0006620 | post-translational protein targeting to endoplasmic reticulum membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0010498 | proteasomal protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0043161 | proteasome-mediated ubiquitin-dependent protein catabolic process | IMP | J:233169 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | IMP | J:139505 | |||||||||
| Biological Process | GO:0050821 | protein stabilization | IMP | J:131590 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | IMP | J:119857 | |||||||||
| Biological Process | GO:0042981 | regulation of apoptotic process | RCA | J:84168 | |||||||||
| Biological Process | GO:0045995 | regulation of embryonic development | IMP | J:119857 | |||||||||
| Biological Process | GO:0031647 | regulation of protein stability | ISO | J:164563 | |||||||||
| Biological Process | GO:0007283 | spermatogenesis | IMP | J:139505 | |||||||||
| Biological Process | GO:0007130 | synaptonemal complex assembly | IMP | J:139505 | |||||||||
| Biological Process | GO:0071816 | tail-anchored membrane protein insertion into ER membrane | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0030433 | ubiquitin-dependent ERAD pathway | IBA | J:265628 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | ISO | J:164563 | |||||||||
| Biological Process | GO:0006511 | ubiquitin-dependent protein catabolic process | IMP | J:168161 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||