|
Symbol Name ID |
Rnf135
ring finger protein 135 MGI:1919206 |
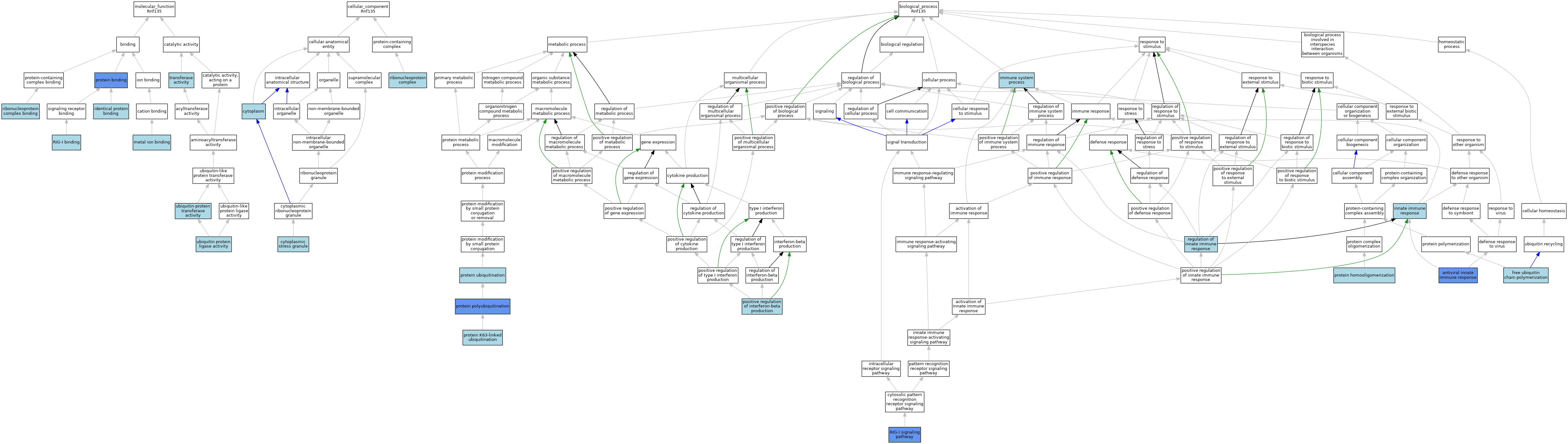
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0042802 | identical protein binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0046872 | metal ion binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:103507 | |||||||||
| Molecular Function | GO:0043021 | ribonucleoprotein complex binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0043021 | ribonucleoprotein complex binding | IBA | J:265628 | |||||||||
| Molecular Function | GO:0039552 | RIG-I binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0016740 | transferase activity | IEA | J:60000 | |||||||||
| Molecular Function | GO:0061630 | ubiquitin protein ligase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004842 | ubiquitin-protein transferase activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0004842 | ubiquitin-protein transferase activity | IBA | J:265628 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IC | J:281202 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0010494 | cytoplasmic stress granule | ISO | J:164563 | |||||||||
| Cellular Component | GO:1990904 | ribonucleoprotein complex | ISO | J:164563 | |||||||||
| Biological Process | GO:0140374 | antiviral innate immune response | IMP | J:261333 | |||||||||
| Biological Process | GO:0140374 | antiviral innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0140374 | antiviral innate immune response | IMP | J:260659 | |||||||||
| Biological Process | GO:0010994 | free ubiquitin chain polymerization | ISO | J:164563 | |||||||||
| Biological Process | GO:0002376 | immune system process | IEA | J:60000 | |||||||||
| Biological Process | GO:0045087 | innate immune response | IEA | J:60000 | |||||||||
| Biological Process | GO:0032728 | positive regulation of interferon-beta production | ISO | J:164563 | |||||||||
| Biological Process | GO:0051260 | protein homooligomerization | ISO | J:164563 | |||||||||
| Biological Process | GO:0070534 | protein K63-linked ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0000209 | protein polyubiquitination | IGI | J:260659 | |||||||||
| Biological Process | GO:0016567 | protein ubiquitination | ISO | J:164563 | |||||||||
| Biological Process | GO:0045088 | regulation of innate immune response | ISO | J:164563 | |||||||||
| Biological Process | GO:0045088 | regulation of innate immune response | IBA | J:265628 | |||||||||
| Biological Process | GO:0039529 | RIG-I signaling pathway | ISO | J:164563 | |||||||||
| Biological Process | GO:0039529 | RIG-I signaling pathway | IMP | J:260659 | |||||||||
| Biological Process | GO:0039529 | RIG-I signaling pathway | IMP | J:281202 | |||||||||
| Biological Process | GO:0039529 | RIG-I signaling pathway | IGI | J:260659 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||