|
Symbol Name ID |
Cic
capicua transcriptional repressor MGI:1918972 |
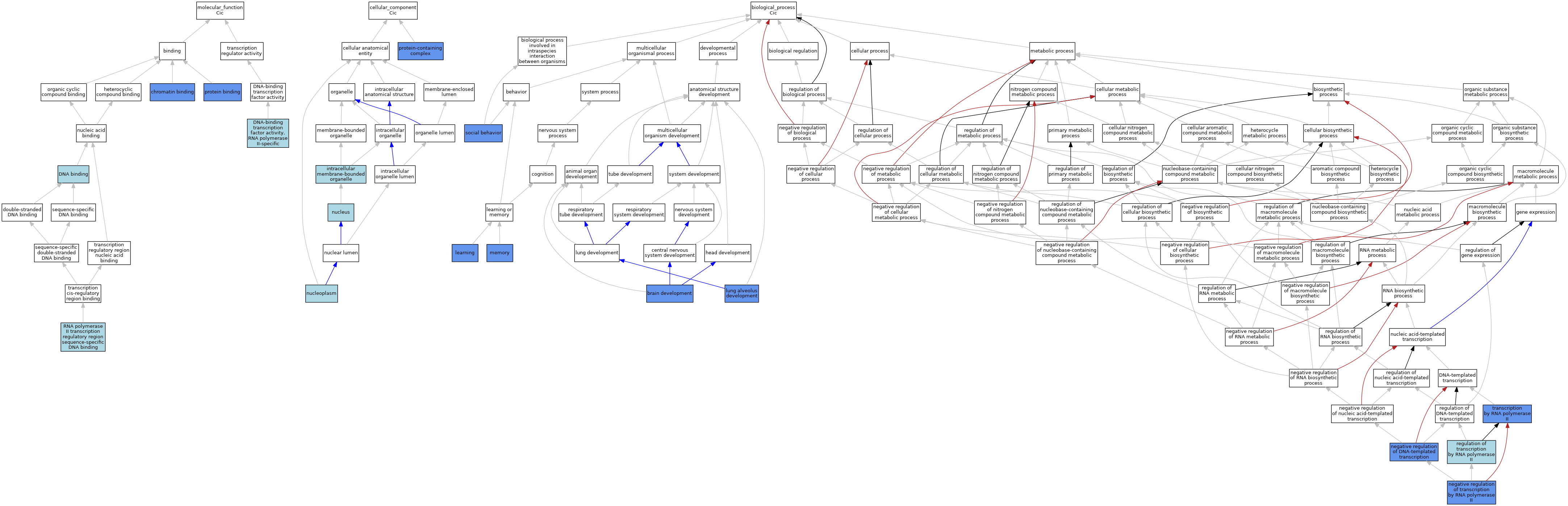
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:162235 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:178303 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:162235 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:178303 | |||||||||
| Molecular Function | GO:0003682 | chromatin binding | IDA | J:178303 | |||||||||
| Molecular Function | GO:0003677 | DNA binding | IEA | J:60000 | |||||||||
| Molecular Function | GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | IBA | J:265628 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:240674 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162235 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:133952 | |||||||||
| Molecular Function | GO:0005515 | protein binding | IPI | J:162235 | |||||||||
| Molecular Function | GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043231 | intracellular membrane-bounded organelle | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0032991 | protein-containing complex | IDA | J:133952 | |||||||||
| Biological Process | GO:0007420 | brain development | IMP | J:240674 | |||||||||
| Biological Process | GO:0007612 | learning | IMP | J:240674 | |||||||||
| Biological Process | GO:0048286 | lung alveolus development | IGI | J:178303 | |||||||||
| Biological Process | GO:0048286 | lung alveolus development | IGI | J:178303 | |||||||||
| Biological Process | GO:0048286 | lung alveolus development | IMP | J:178303 | |||||||||
| Biological Process | GO:0007613 | memory | IMP | J:240674 | |||||||||
| Biological Process | GO:0045892 | negative regulation of DNA-templated transcription | IMP | J:240674 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:178303 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:162235 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | IGI | J:178303 | |||||||||
| Biological Process | GO:0006357 | regulation of transcription by RNA polymerase II | IBA | J:265628 | |||||||||
| Biological Process | GO:0035176 | social behavior | IMP | J:240674 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:178303 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:178303 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:178303 | |||||||||
| Biological Process | GO:0006366 | transcription by RNA polymerase II | IGI | J:178303 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/23/2024 MGI 6.23 |

|
|
|
||