|
Symbol Name ID |
Zmynd8
zinc finger, MYND-type containing 8 MGI:1918025 |
| Category | GO ID | Classification Term | Evidence | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
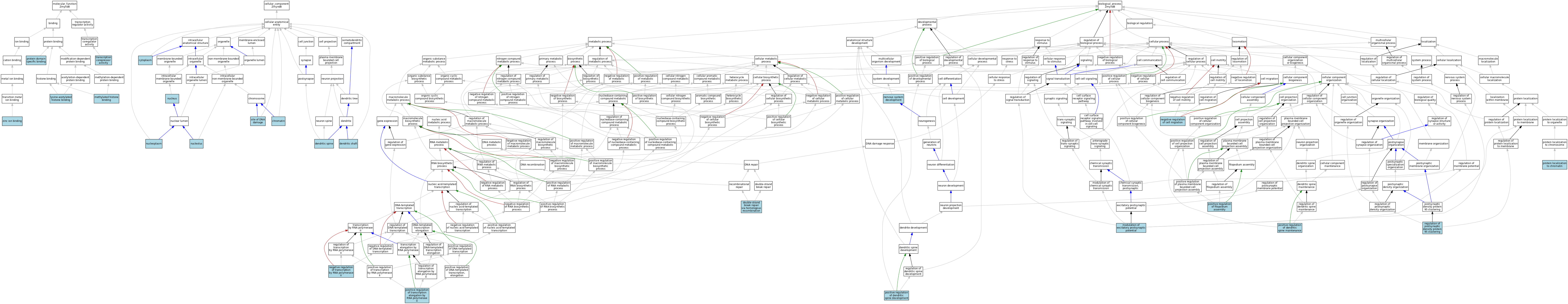
| Molecular Function | GO:0070577 | lysine-acetylated histone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0035064 | methylated histone binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0019904 | protein domain specific binding | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | ISO | J:164563 | |||||||||
| Molecular Function | GO:0003714 | transcription corepressor activity | IBA | J:265628 | |||||||||
| Molecular Function | GO:0008270 | zinc ion binding | ISO | J:164563 | |||||||||
| Cellular Component | GO:0000785 | chromatin | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005737 | cytoplasm | IBA | J:265628 | |||||||||
| Cellular Component | GO:0043198 | dendritic shaft | ISO | J:155856 | |||||||||
| Cellular Component | GO:0043197 | dendritic spine | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005730 | nucleolus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005654 | nucleoplasm | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:155856 | |||||||||
| Cellular Component | GO:0005634 | nucleus | ISO | J:164563 | |||||||||
| Cellular Component | GO:0005634 | nucleus | IBA | J:265628 | |||||||||
| Cellular Component | GO:0090734 | site of DNA damage | ISO | J:164563 | |||||||||
| Biological Process | GO:0000724 | double-strand break repair via homologous recombination | ISO | J:164563 | |||||||||
| Biological Process | GO:0098815 | modulation of excitatory postsynaptic potential | ISO | J:155856 | |||||||||
| Biological Process | GO:0030336 | negative regulation of cell migration | ISO | J:164563 | |||||||||
| Biological Process | GO:0000122 | negative regulation of transcription by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0007399 | nervous system development | ISO | J:164563 | |||||||||
| Biological Process | GO:0060999 | positive regulation of dendritic spine development | ISO | J:155856 | |||||||||
| Biological Process | GO:1902952 | positive regulation of dendritic spine maintenance | ISO | J:155856 | |||||||||
| Biological Process | GO:0051491 | positive regulation of filopodium assembly | ISO | J:155856 | |||||||||
| Biological Process | GO:0032968 | positive regulation of transcription elongation by RNA polymerase II | ISO | J:164563 | |||||||||
| Biological Process | GO:0071168 | protein localization to chromatin | ISO | J:164563 | |||||||||
| Biological Process | GO:1902897 | regulation of postsynaptic density protein 95 clustering | ISO | J:155856 | |||||||||
Mouse Genome Database (MGD), Gene Expression Database (GXD), Mouse Models of Human Cancer database (MMHCdb) (formerly Mouse Tumor Biology (MTB)), Gene Ontology (GO) |
||
|
Citing These Resources Funding Information Warranty Disclaimer, Privacy Notice, Licensing, & Copyright Send questions and comments to User Support. |
last database update 04/16/2024 MGI 6.23 |

|
|
|
||